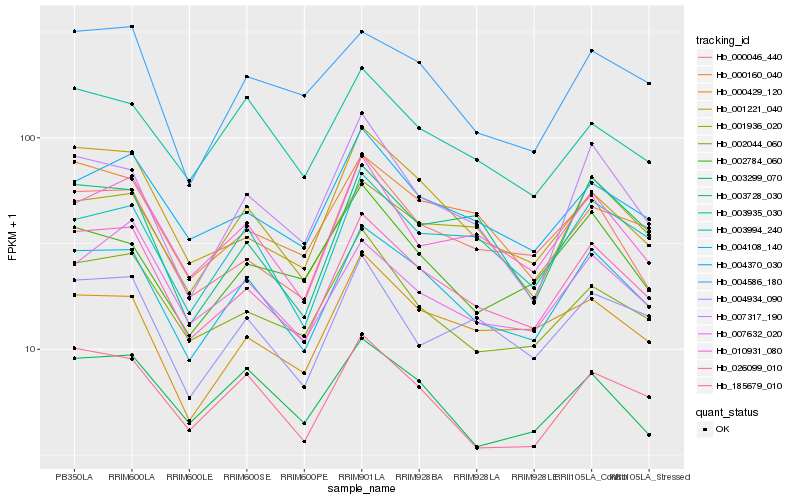
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004108_140 |
0.0 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 2 |
Hb_026099_010 |
0.0460131161 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 3 |
Hb_185679_010 |
0.0659749756 |
- |
- |
serine/threonine-protein kinase rio2, putative [Ricinus communis] |
| 4 |
Hb_003994_240 |
0.0701412236 |
- |
- |
Mediator subunit 8 isoform 3 [Theobroma cacao] |
| 5 |
Hb_001936_020 |
0.0725494197 |
- |
- |
PREDICTED: phosphatidylinositol 3-kinase, root isoform isoform X1 [Jatropha curcas] |
| 6 |
Hb_003935_030 |
0.0790098134 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 kappa isoform X2 [Jatropha curcas] |
| 7 |
Hb_004370_030 |
0.0792541756 |
- |
- |
PREDICTED: RNA-binding protein 34 [Jatropha curcas] |
| 8 |
Hb_003299_070 |
0.0811997434 |
- |
- |
AMP deaminase [Theobroma cacao] |
| 9 |
Hb_007317_190 |
0.0817976927 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI1 [Jatropha curcas] |
| 10 |
Hb_004586_180 |
0.0819342784 |
- |
- |
hypothetical protein JCGZ_15866 [Jatropha curcas] |
| 11 |
Hb_010931_080 |
0.0821011137 |
- |
- |
PREDICTED: U4/U6.U5 tri-snRNP-associated protein 2-like [Jatropha curcas] |
| 12 |
Hb_002044_060 |
0.0840685182 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 13 |
Hb_000046_440 |
0.0859266107 |
transcription factor |
TF Family: SNF2 |
PREDICTED: putative chromatin-remodeling complex ATPase chain isoform X1 [Jatropha curcas] |
| 14 |
Hb_007632_020 |
0.0865813405 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 17 [Jatropha curcas] |
| 15 |
Hb_002784_060 |
0.0870315648 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA6 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000160_040 |
0.08713616 |
- |
- |
PREDICTED: methyltransferase-like protein 10 [Jatropha curcas] |
| 17 |
Hb_003728_030 |
0.0883003572 |
- |
- |
o-sialoglycoprotein endopeptidase, putative [Ricinus communis] |
| 18 |
Hb_000429_120 |
0.0894581983 |
- |
- |
PREDICTED: DNA-directed RNA polymerase I subunit rpa1 [Jatropha curcas] |
| 19 |
Hb_001221_040 |
0.091269889 |
- |
- |
PREDICTED: nucleolar protein 56-like [Jatropha curcas] |
| 20 |
Hb_004934_090 |
0.0915967065 |
- |
- |
hypothetical protein JCGZ_08073 [Jatropha curcas] |