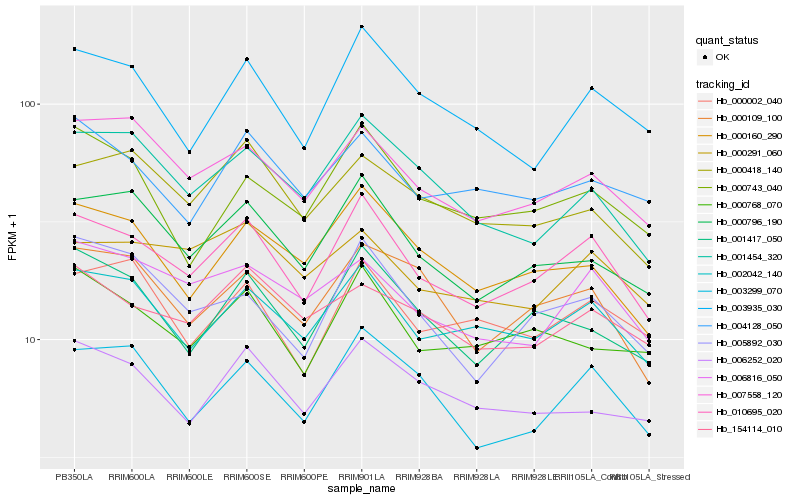
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010695_020 |
0.0 |
- |
- |
PREDICTED: superkiller viralicidic activity 2-like 2 [Jatropha curcas] |
| 2 |
Hb_002042_140 |
0.0621727089 |
- |
- |
PREDICTED: uncharacterized protein LOC105647570 [Jatropha curcas] |
| 3 |
Hb_000796_190 |
0.0716402236 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 4 |
Hb_007558_120 |
0.0740413393 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 5 |
Hb_003299_070 |
0.0740784981 |
- |
- |
AMP deaminase [Theobroma cacao] |
| 6 |
Hb_000160_290 |
0.079012448 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_003935_030 |
0.0793401175 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 kappa isoform X2 [Jatropha curcas] |
| 8 |
Hb_001417_050 |
0.0800839285 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 9 |
Hb_000768_070 |
0.0826171372 |
transcription factor |
TF Family: SNF2 |
PREDICTED: TATA-binding protein-associated factor BTAF1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_005892_030 |
0.0843877961 |
- |
- |
PREDICTED: splicing factor U2af large subunit A [Jatropha curcas] |
| 11 |
Hb_000109_100 |
0.0847115992 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 [Jatropha curcas] |
| 12 |
Hb_006816_050 |
0.085266663 |
- |
- |
PREDICTED: RNA polymerase II-associated protein 3 isoform X4 [Jatropha curcas] |
| 13 |
Hb_000002_040 |
0.0858600838 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 14 |
Hb_001454_320 |
0.0860123647 |
- |
- |
PREDICTED: protein pleiotropic regulatory locus 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000291_060 |
0.0879470278 |
transcription factor |
TF Family: C2H2 |
PREDICTED: MYST-like histone acetyltransferase 2 [Jatropha curcas] |
| 16 |
Hb_000418_140 |
0.0882660614 |
- |
- |
PREDICTED: DNA damage-binding protein 1a [Jatropha curcas] |
| 17 |
Hb_004128_050 |
0.0884505699 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_006252_020 |
0.0886748586 |
- |
- |
PREDICTED: SET and MYND domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 19 |
Hb_154114_010 |
0.0896864848 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3A isoform X2 [Jatropha curcas] |
| 20 |
Hb_000743_040 |
0.0901406371 |
- |
- |
ribosomal RNA methyltransferase, putative [Ricinus communis] |