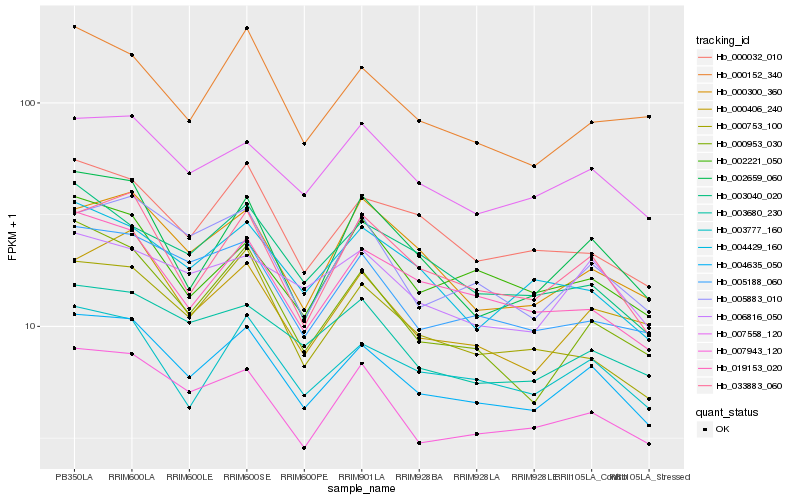
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004635_050 |
0.0 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 2 |
Hb_007943_120 |
0.0605076141 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 5 isoform X4 [Jatropha curcas] |
| 3 |
Hb_007558_120 |
0.0631785466 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 4 |
Hb_033883_060 |
0.0644870615 |
- |
- |
PREDICTED: uncharacterized protein LOC105648353 [Jatropha curcas] |
| 5 |
Hb_003777_160 |
0.0647540626 |
transcription factor |
TF Family: bZIP |
PREDICTED: G-box-binding factor 4 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005883_010 |
0.0698418203 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_019153_020 |
0.0708872353 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003680_230 |
0.0732165003 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 9 |
Hb_000032_010 |
0.0735705756 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 10 |
Hb_005188_060 |
0.074036407 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 11 |
Hb_000406_240 |
0.0747000441 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Jatropha curcas] |
| 12 |
Hb_002659_060 |
0.0785591276 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3B [Jatropha curcas] |
| 13 |
Hb_000300_360 |
0.0787531596 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 14 |
Hb_000753_100 |
0.0794190685 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640677 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004429_160 |
0.0796284169 |
- |
- |
PREDICTED: serine/threonine-protein kinase TAO3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003040_020 |
0.0800207966 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 17 |
Hb_002221_050 |
0.0801923012 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP65 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000152_340 |
0.0818880074 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 19 |
Hb_006816_050 |
0.0819245332 |
- |
- |
PREDICTED: RNA polymerase II-associated protein 3 isoform X4 [Jatropha curcas] |
| 20 |
Hb_000953_030 |
0.0824156836 |
transcription factor |
TF Family: HSF |
DNA binding protein, putative [Ricinus communis] |