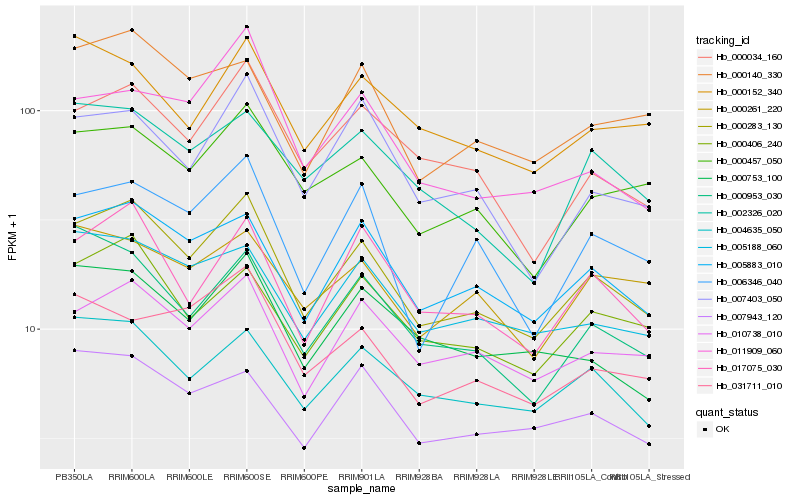
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000283_130 |
0.0 |
- |
- |
PREDICTED: PRA1 family protein E-like [Populus euphratica] |
| 2 |
Hb_005883_010 |
0.0651311893 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_010738_010 |
0.0767489115 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 4 |
Hb_000406_240 |
0.0795433618 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Jatropha curcas] |
| 5 |
Hb_017075_030 |
0.0796543408 |
- |
- |
helicase, putative [Ricinus communis] |
| 6 |
Hb_000457_050 |
0.0813974612 |
- |
- |
PREDICTED: tether containing UBX domain for GLUT4 [Jatropha curcas] |
| 7 |
Hb_004635_050 |
0.0839649209 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 8 |
Hb_000034_160 |
0.0856771999 |
- |
- |
hypothetical protein EUGRSUZ_F02151 [Eucalyptus grandis] |
| 9 |
Hb_005188_060 |
0.0867537272 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 10 |
Hb_000140_330 |
0.0875626218 |
- |
- |
PREDICTED: uncharacterized protein LOC105642909 [Jatropha curcas] |
| 11 |
Hb_006346_040 |
0.0923715441 |
- |
- |
hypothetical protein JCGZ_20335 [Jatropha curcas] |
| 12 |
Hb_007943_120 |
0.0926061286 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 5 isoform X4 [Jatropha curcas] |
| 13 |
Hb_007403_050 |
0.0928200488 |
- |
- |
PREDICTED: protein YIPF5 homolog [Jatropha curcas] |
| 14 |
Hb_002326_020 |
0.0941548913 |
- |
- |
hypothetical protein POPTR_0002s13160g [Populus trichocarpa] |
| 15 |
Hb_000753_100 |
0.0942484639 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640677 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000953_030 |
0.0946995372 |
transcription factor |
TF Family: HSF |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_031711_010 |
0.0970312095 |
- |
- |
PREDICTED: CDPK-related kinase 4-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000152_340 |
0.0984486452 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 19 |
Hb_000261_220 |
0.1013550136 |
- |
- |
PREDICTED: TBC1 domain family member 13 [Jatropha curcas] |
| 20 |
Hb_011909_060 |
0.1027658892 |
- |
- |
PREDICTED: zinc finger protein CONSTANS-LIKE 1 [Jatropha curcas] |