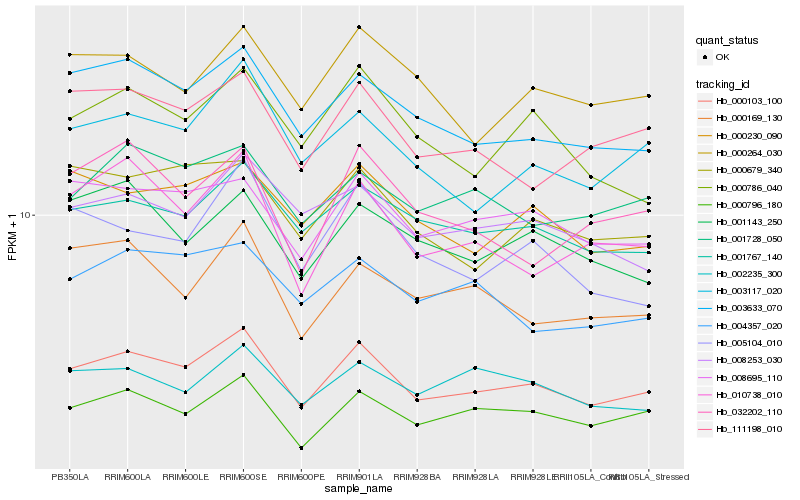
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000103_100 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g22410, mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_003633_070 |
0.0689977643 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 3 |
Hb_008695_110 |
0.0739493037 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105638874 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000796_180 |
0.0751110933 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Vitis vinifera] |
| 5 |
Hb_003117_020 |
0.0755495662 |
transcription factor |
TF Family: BBR-BPC |
PREDICTED: protein BASIC PENTACYSTEINE6 [Jatropha curcas] |
| 6 |
Hb_010738_010 |
0.0760654996 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 7 |
Hb_001767_140 |
0.0775906018 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 8 |
Hb_111198_010 |
0.0789409859 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000230_090 |
0.0827646981 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000786_040 |
0.083111219 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 17 [Jatropha curcas] |
| 11 |
Hb_000679_340 |
0.0843295525 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005104_010 |
0.0851453867 |
- |
- |
PREDICTED: uncharacterized protein LOC105140796 isoform X1 [Populus euphratica] |
| 13 |
Hb_000169_130 |
0.0872305936 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 14 |
Hb_008253_030 |
0.0875411214 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 15 |
Hb_001143_250 |
0.0877237156 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3C isoform X1 [Jatropha curcas] |
| 16 |
Hb_000264_030 |
0.0879598129 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |
| 17 |
Hb_032202_110 |
0.0887862017 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 18 |
Hb_004357_020 |
0.0891251897 |
- |
- |
PREDICTED: uncharacterized protein LOC105637355 [Jatropha curcas] |
| 19 |
Hb_001728_050 |
0.0891346382 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |
| 20 |
Hb_002235_300 |
0.0902299663 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15280 isoform X1 [Jatropha curcas] |