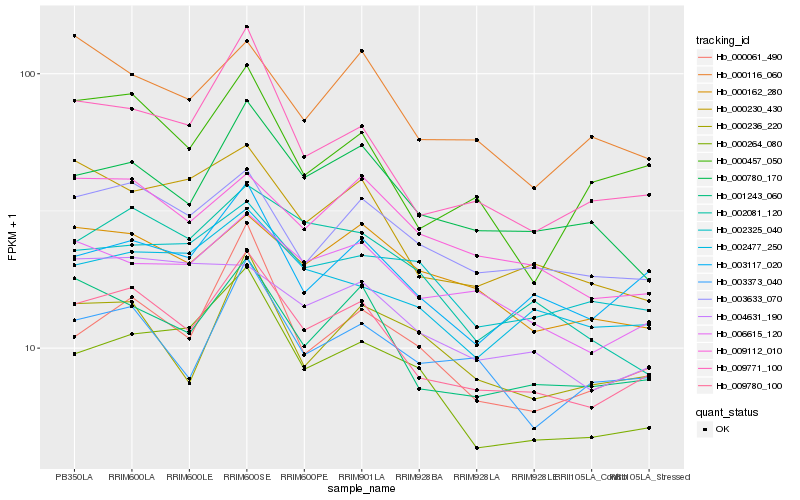
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009780_100 |
0.0 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 2 |
Hb_001243_060 |
0.0580869465 |
- |
- |
PREDICTED: probable magnesium transporter NIPA6 [Jatropha curcas] |
| 3 |
Hb_009771_100 |
0.0710368559 |
- |
- |
PREDICTED: uncharacterized protein LOC105141784 [Populus euphratica] |
| 4 |
Hb_003633_070 |
0.076396047 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 5 |
Hb_006615_120 |
0.0769838547 |
- |
- |
PREDICTED: uncharacterized protein LOC105640682 [Jatropha curcas] |
| 6 |
Hb_003117_020 |
0.0774499672 |
transcription factor |
TF Family: BBR-BPC |
PREDICTED: protein BASIC PENTACYSTEINE6 [Jatropha curcas] |
| 7 |
Hb_000230_430 |
0.0776768133 |
- |
- |
catalytic, putative [Ricinus communis] |
| 8 |
Hb_000780_170 |
0.0785245497 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 9 |
Hb_009112_010 |
0.0801504675 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 10 |
Hb_003373_040 |
0.0817552949 |
- |
- |
hypothetical protein ARALYDRAFT_356474 [Arabidopsis lyrata subsp. lyrata] |
| 11 |
Hb_000061_490 |
0.0819155261 |
- |
- |
hypothetical protein CISIN_1g0362002mg, partial [Citrus sinensis] |
| 12 |
Hb_002477_250 |
0.0822268217 |
- |
- |
DNA-directed RNA polymerase I 49 kDa polypeptide, putative [Ricinus communis] |
| 13 |
Hb_000162_280 |
0.0832197996 |
- |
- |
PREDICTED: CTL-like protein DDB_G0288717 [Populus euphratica] |
| 14 |
Hb_000264_080 |
0.0860844485 |
- |
- |
hypothetical protein JCGZ_21412 [Jatropha curcas] |
| 15 |
Hb_000457_050 |
0.0885296659 |
- |
- |
PREDICTED: tether containing UBX domain for GLUT4 [Jatropha curcas] |
| 16 |
Hb_000116_060 |
0.0887718672 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 17 |
Hb_000236_220 |
0.0892580412 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 18 |
Hb_002081_120 |
0.089890932 |
- |
- |
PREDICTED: magnesium transporter MRS2-3 [Jatropha curcas] |
| 19 |
Hb_004631_190 |
0.0899420212 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 20 |
Hb_002325_040 |
0.0903756622 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |