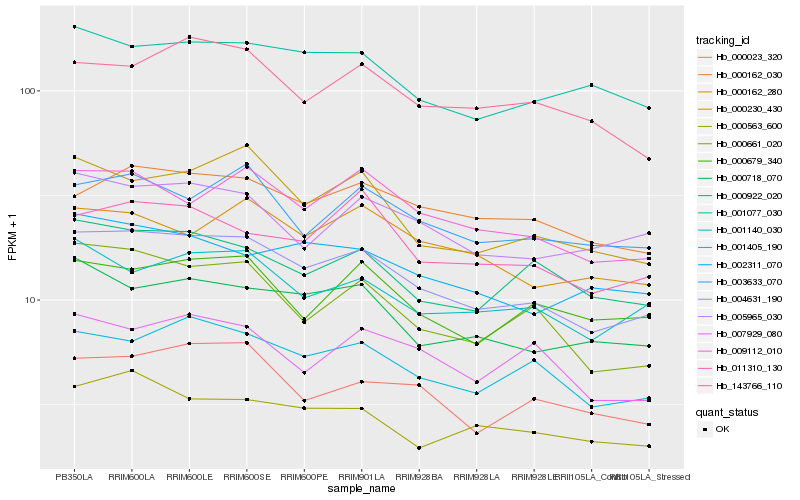
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004631_190 |
0.0 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 2 |
Hb_001405_190 |
0.0608862336 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 3 |
Hb_009112_010 |
0.0684909499 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 4 |
Hb_000230_430 |
0.0689490749 |
- |
- |
catalytic, putative [Ricinus communis] |
| 5 |
Hb_001140_030 |
0.0699857195 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase XBAT33 [Jatropha curcas] |
| 6 |
Hb_000718_070 |
0.0699906408 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000661_020 |
0.0747685552 |
- |
- |
BnaC08g27190D [Brassica napus] |
| 8 |
Hb_003633_070 |
0.0750385079 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 9 |
Hb_001077_030 |
0.0758325412 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_011310_130 |
0.0767144397 |
- |
- |
DEAD/DEAH box RNA helicase family protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_000922_020 |
0.0767294027 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 12 |
Hb_000162_030 |
0.0768312701 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_005965_030 |
0.076959815 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |
| 14 |
Hb_143766_110 |
0.0775960503 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 1 [Jatropha curcas] |
| 15 |
Hb_000563_600 |
0.0782799843 |
- |
- |
PREDICTED: uncharacterized protein LOC104428079 [Eucalyptus grandis] |
| 16 |
Hb_000679_340 |
0.0786532238 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002311_070 |
0.0790896062 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 18 |
Hb_000023_320 |
0.0794186304 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000162_280 |
0.0812278588 |
- |
- |
PREDICTED: CTL-like protein DDB_G0288717 [Populus euphratica] |
| 20 |
Hb_007929_080 |
0.0826290865 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |