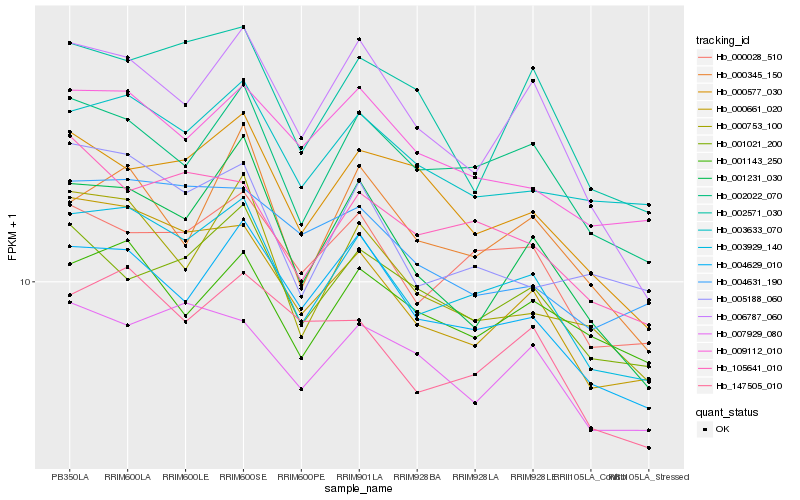
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003929_140 |
0.0 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_000661_020 |
0.0646130366 |
- |
- |
BnaC08g27190D [Brassica napus] |
| 3 |
Hb_002022_070 |
0.0698313839 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 4 |
Hb_000028_510 |
0.0711974137 |
- |
- |
PREDICTED: uncharacterized protein LOC105640819 [Jatropha curcas] |
| 5 |
Hb_004629_010 |
0.0728629167 |
- |
- |
PREDICTED: AP-3 complex subunit delta [Jatropha curcas] |
| 6 |
Hb_000753_100 |
0.079625312 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640677 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001231_030 |
0.0798686275 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |
| 8 |
Hb_147505_010 |
0.080287564 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 9 |
Hb_006787_060 |
0.0836781789 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase SUD1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001021_200 |
0.0845162616 |
desease resistance |
Gene Name: AAA |
PREDICTED: peroxisome biogenesis protein 6 [Jatropha curcas] |
| 11 |
Hb_105641_010 |
0.0861729073 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 12 |
Hb_003633_070 |
0.0866961435 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 13 |
Hb_005188_060 |
0.0869051482 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 14 |
Hb_009112_010 |
0.08878157 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 15 |
Hb_007929_080 |
0.089075083 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |
| 16 |
Hb_000577_030 |
0.0900697457 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase 12 -like protein [Gossypium arboreum] |
| 17 |
Hb_000345_150 |
0.0913594646 |
- |
- |
PREDICTED: uncharacterized protein LOC105647517 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002571_030 |
0.092222325 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |
| 19 |
Hb_004631_190 |
0.0930118387 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 20 |
Hb_001143_250 |
0.0931565179 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3C isoform X1 [Jatropha curcas] |