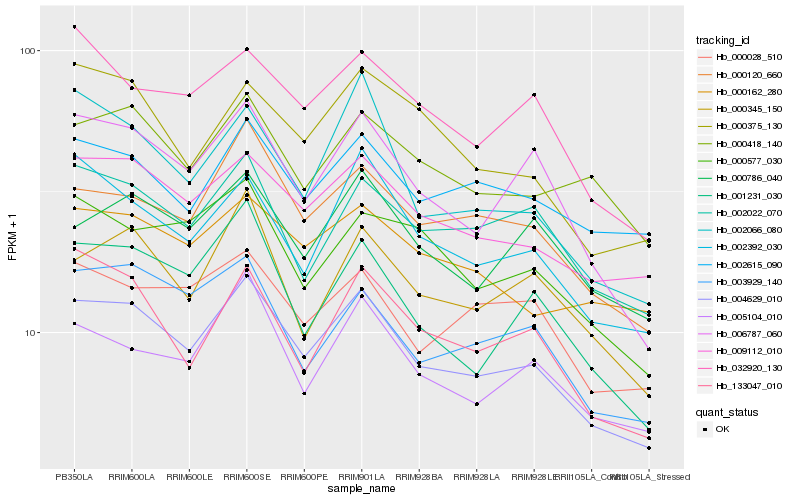
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004629_010 |
0.0 |
- |
- |
PREDICTED: AP-3 complex subunit delta [Jatropha curcas] |
| 2 |
Hb_009112_010 |
0.0551462667 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 3 |
Hb_006787_060 |
0.0661151944 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase SUD1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000120_660 |
0.0663593004 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 5 |
Hb_002392_030 |
0.0679880996 |
- |
- |
WD-40 repeat family protein / small nuclear ribonucleoprotein Prp4p-related [Theobroma cacao] |
| 6 |
Hb_005104_010 |
0.0707820118 |
- |
- |
PREDICTED: uncharacterized protein LOC105140796 isoform X1 [Populus euphratica] |
| 7 |
Hb_003929_140 |
0.0728629167 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_002022_070 |
0.0733420479 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 9 |
Hb_032920_130 |
0.0742894465 |
- |
- |
PREDICTED: vacuolar-sorting receptor 3 [Jatropha curcas] |
| 10 |
Hb_001231_030 |
0.0753152866 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |
| 11 |
Hb_133047_010 |
0.0753712617 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_000345_150 |
0.0765980802 |
- |
- |
PREDICTED: uncharacterized protein LOC105647517 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000375_130 |
0.0771376834 |
- |
- |
Leucine-rich repeat transmembrane protein kinase family protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_000786_040 |
0.0783190294 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 17 [Jatropha curcas] |
| 15 |
Hb_002066_080 |
0.0783446019 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 16 |
Hb_000028_510 |
0.0783448452 |
- |
- |
PREDICTED: uncharacterized protein LOC105640819 [Jatropha curcas] |
| 17 |
Hb_000162_280 |
0.079299333 |
- |
- |
PREDICTED: CTL-like protein DDB_G0288717 [Populus euphratica] |
| 18 |
Hb_000418_140 |
0.079539524 |
- |
- |
PREDICTED: DNA damage-binding protein 1a [Jatropha curcas] |
| 19 |
Hb_002615_090 |
0.0801656843 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000577_030 |
0.0806028596 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase 12 -like protein [Gossypium arboreum] |