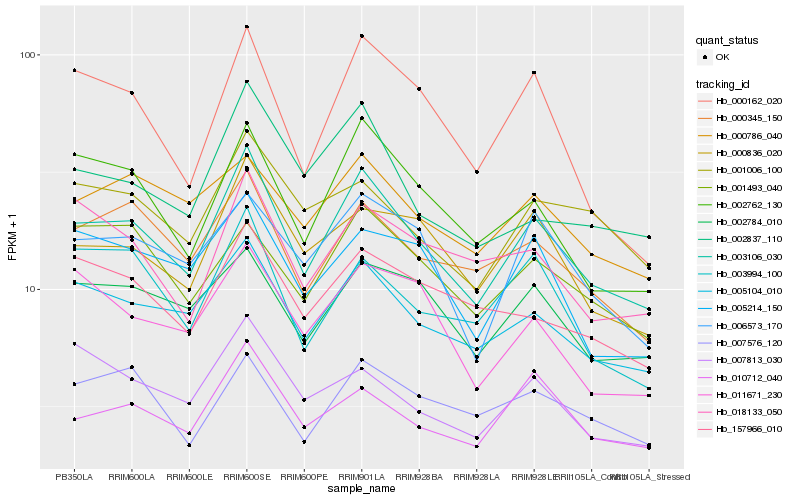
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003106_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 2 |
Hb_005104_010 |
0.0743533577 |
- |
- |
PREDICTED: uncharacterized protein LOC105140796 isoform X1 [Populus euphratica] |
| 3 |
Hb_002762_130 |
0.0875464954 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 1 [Jatropha curcas] |
| 4 |
Hb_000345_150 |
0.0891847676 |
- |
- |
PREDICTED: uncharacterized protein LOC105647517 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000836_020 |
0.0906170438 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 6 |
Hb_010712_040 |
0.0924128311 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g11290 [Jatropha curcas] |
| 7 |
Hb_002784_010 |
0.0941834362 |
- |
- |
DNA-directed RNA polymerase I subunit, putative [Ricinus communis] |
| 8 |
Hb_006573_170 |
0.0985253822 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002837_110 |
0.100042506 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 2, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000162_020 |
0.1007919637 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 11 |
Hb_000786_040 |
0.1012367227 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 17 [Jatropha curcas] |
| 12 |
Hb_005214_150 |
0.1014624176 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 13 |
Hb_007576_120 |
0.103221805 |
- |
- |
NBS-LRR disease resistance protein NBS49 [Dimocarpus longan] |
| 14 |
Hb_157966_010 |
0.1043331583 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 15 |
Hb_001493_040 |
0.10443514 |
- |
- |
PREDICTED: FACT complex subunit SPT16-like [Jatropha curcas] |
| 16 |
Hb_003994_100 |
0.1047685738 |
transcription factor |
TF Family: PHD |
PREDICTED: histone acetyltransferase HAC1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001006_100 |
0.1054104416 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: SWI/SNF complex component SNF12 homolog isoform X1 [Jatropha curcas] |
| 18 |
Hb_011671_230 |
0.105590878 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_018133_050 |
0.1063011968 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 1 [Jatropha curcas] |
| 20 |
Hb_007813_030 |
0.1064008198 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At4g19050 [Jatropha curcas] |