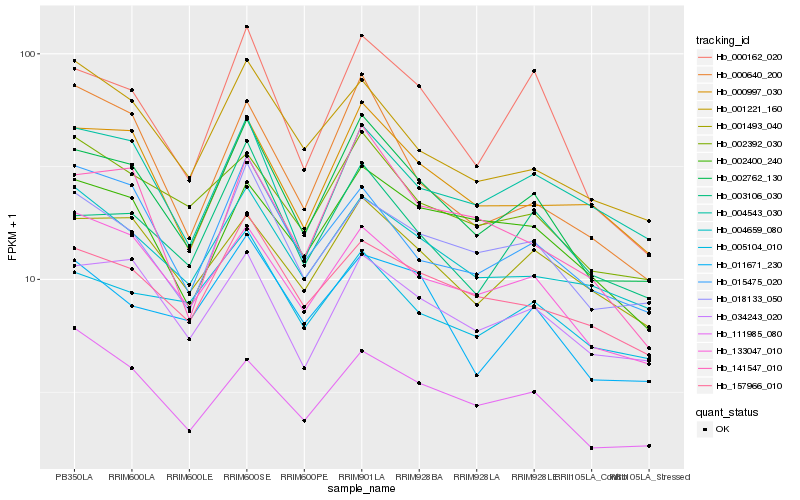
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002762_130 |
0.0 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 1 [Jatropha curcas] |
| 2 |
Hb_000162_020 |
0.0760948141 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 3 |
Hb_018133_050 |
0.0812023385 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 1 [Jatropha curcas] |
| 4 |
Hb_034243_020 |
0.0815254598 |
- |
- |
PREDICTED: elongator complex protein 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000997_030 |
0.0835059654 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 6 |
Hb_133047_010 |
0.0858244671 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_001493_040 |
0.0861587488 |
- |
- |
PREDICTED: FACT complex subunit SPT16-like [Jatropha curcas] |
| 8 |
Hb_003106_030 |
0.0875464954 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 9 |
Hb_157966_010 |
0.0884170099 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 10 |
Hb_002392_030 |
0.090161661 |
- |
- |
WD-40 repeat family protein / small nuclear ribonucleoprotein Prp4p-related [Theobroma cacao] |
| 11 |
Hb_004659_080 |
0.090862899 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 12 |
Hb_011671_230 |
0.0912646462 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001221_160 |
0.0960198215 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 14 |
Hb_002400_240 |
0.0963505172 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 15 |
Hb_141547_010 |
0.096935873 |
- |
- |
PREDICTED: ARF guanine-nucleotide exchange factor GNOM [Jatropha curcas] |
| 16 |
Hb_004543_030 |
0.0981567698 |
- |
- |
PREDICTED: FIP1[V]-like protein [Jatropha curcas] |
| 17 |
Hb_015475_020 |
0.0982335958 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 18 |
Hb_005104_010 |
0.0984255415 |
- |
- |
PREDICTED: uncharacterized protein LOC105140796 isoform X1 [Populus euphratica] |
| 19 |
Hb_111985_080 |
0.0996427344 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g19720 [Jatropha curcas] |
| 20 |
Hb_000640_200 |
0.1020985734 |
- |
- |
hypothetical protein RCOM_1346600 [Ricinus communis] |