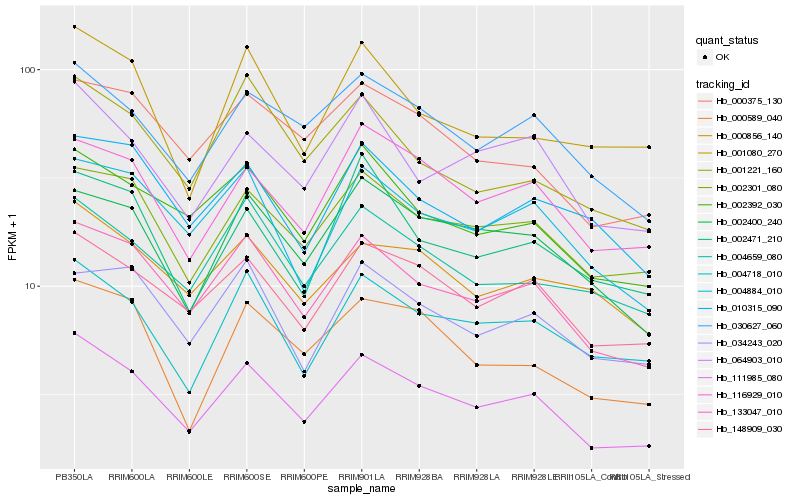
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_111985_080 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g19720 [Jatropha curcas] |
| 2 |
Hb_133047_010 |
0.0513807095 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_148909_030 |
0.0691024678 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002301_080 |
0.0729971082 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X2 [Jatropha curcas] |
| 5 |
Hb_030627_060 |
0.0775934306 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 6 |
Hb_004718_010 |
0.0804554158 |
- |
- |
PREDICTED: uncharacterized protein LOC105638290 [Jatropha curcas] |
| 7 |
Hb_064903_010 |
0.0811792398 |
- |
- |
PREDICTED: T-complex protein 1 subunit delta [Jatropha curcas] |
| 8 |
Hb_002392_030 |
0.0813905067 |
- |
- |
WD-40 repeat family protein / small nuclear ribonucleoprotein Prp4p-related [Theobroma cacao] |
| 9 |
Hb_116929_010 |
0.0826821128 |
- |
- |
PREDICTED: nuclear-pore anchor isoform X2 [Jatropha curcas] |
| 10 |
Hb_004884_010 |
0.0843152255 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC22 homolog [Jatropha curcas] |
| 11 |
Hb_002400_240 |
0.0858147239 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 12 |
Hb_000375_130 |
0.0872717152 |
- |
- |
Leucine-rich repeat transmembrane protein kinase family protein isoform 1 [Theobroma cacao] |
| 13 |
Hb_004659_080 |
0.0898234641 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 14 |
Hb_000589_040 |
0.0898524275 |
desease resistance |
Gene Name: zf-BED |
PREDICTED: probable disease resistance protein At1g12280 [Jatropha curcas] |
| 15 |
Hb_000856_140 |
0.0938188471 |
- |
- |
PREDICTED: uncharacterized protein LOC105640478 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001080_270 |
0.0939248955 |
- |
- |
PREDICTED: NF-kappa-B-activating protein [Jatropha curcas] |
| 17 |
Hb_002471_210 |
0.0940145001 |
- |
- |
PREDICTED: THO complex subunit 2 [Jatropha curcas] |
| 18 |
Hb_001221_160 |
0.0950371186 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 19 |
Hb_010315_090 |
0.0954747599 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 20 |
Hb_034243_020 |
0.0958766054 |
- |
- |
PREDICTED: elongator complex protein 1 isoform X1 [Jatropha curcas] |