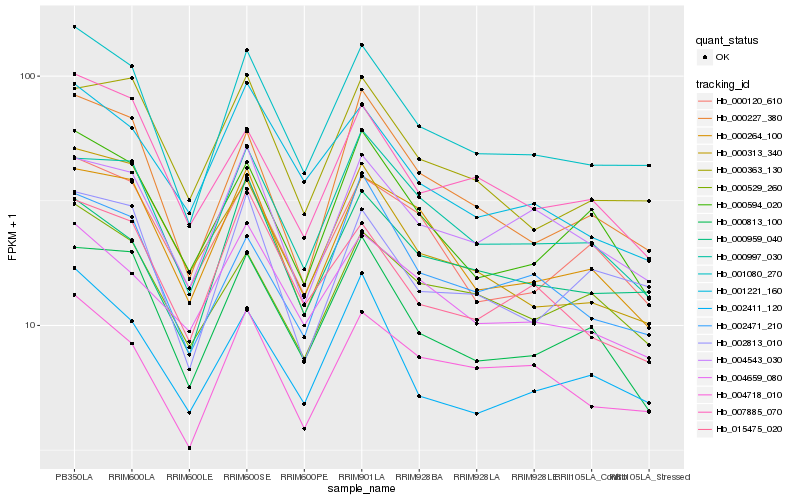
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001080_270 |
0.0 |
- |
- |
PREDICTED: NF-kappa-B-activating protein [Jatropha curcas] |
| 2 |
Hb_000227_380 |
0.0724337409 |
- |
- |
PREDICTED: protein TIME FOR COFFEE isoform X3 [Jatropha curcas] |
| 3 |
Hb_004718_010 |
0.0747494609 |
- |
- |
PREDICTED: uncharacterized protein LOC105638290 [Jatropha curcas] |
| 4 |
Hb_002411_120 |
0.0754062673 |
- |
- |
PREDICTED: uncharacterized protein LOC105631645 [Jatropha curcas] |
| 5 |
Hb_000997_030 |
0.0774018339 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 6 |
Hb_000959_040 |
0.0777701249 |
- |
- |
PREDICTED: uncharacterized protein LOC105636570 [Jatropha curcas] |
| 7 |
Hb_000813_100 |
0.078577775 |
- |
- |
PREDICTED: uncharacterized protein LOC105631583 isoform X2 [Jatropha curcas] |
| 8 |
Hb_002813_010 |
0.0792623216 |
- |
- |
PREDICTED: uncharacterized protein LOC105641844 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002471_210 |
0.0813486476 |
- |
- |
PREDICTED: THO complex subunit 2 [Jatropha curcas] |
| 10 |
Hb_000529_260 |
0.082438129 |
- |
- |
PREDICTED: protein SGT1 homolog At5g65490 [Jatropha curcas] |
| 11 |
Hb_007885_070 |
0.0826321359 |
- |
- |
cleavage and polyadenylation specificity factor, putative [Ricinus communis] |
| 12 |
Hb_000363_130 |
0.0831686994 |
- |
- |
PREDICTED: FRIGIDA-like protein 1 [Jatropha curcas] |
| 13 |
Hb_015475_020 |
0.0833448663 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 14 |
Hb_000264_100 |
0.0834233842 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000120_610 |
0.0846829196 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 16 |
Hb_004543_030 |
0.0847369808 |
- |
- |
PREDICTED: FIP1[V]-like protein [Jatropha curcas] |
| 17 |
Hb_004659_080 |
0.0852256297 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 18 |
Hb_000313_340 |
0.0863098221 |
- |
- |
DNA repair protein xp-E, putative [Ricinus communis] |
| 19 |
Hb_001221_160 |
0.0866688816 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 20 |
Hb_000594_020 |
0.0868258074 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25 isoform X2 [Jatropha curcas] |