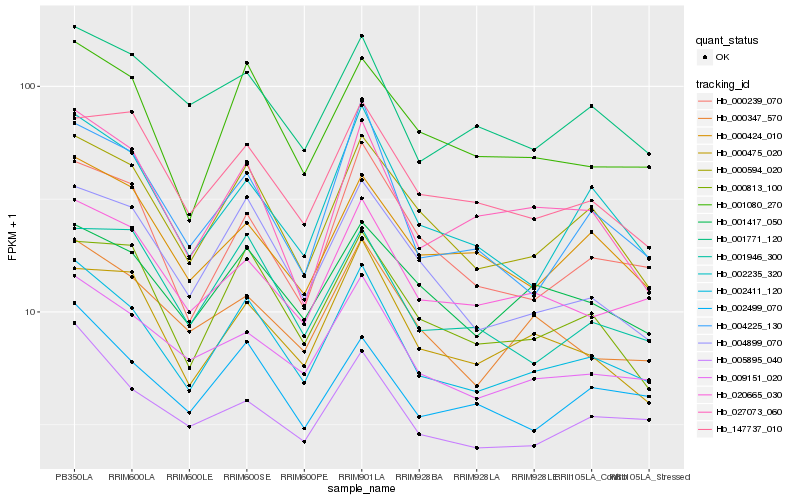
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002411_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631645 [Jatropha curcas] |
| 2 |
Hb_000594_020 |
0.0702008782 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25 isoform X2 [Jatropha curcas] |
| 3 |
Hb_009151_020 |
0.0724825604 |
- |
- |
PREDICTED: DNA polymerase delta catalytic subunit [Jatropha curcas] |
| 4 |
Hb_001080_270 |
0.0754062673 |
- |
- |
PREDICTED: NF-kappa-B-activating protein [Jatropha curcas] |
| 5 |
Hb_020665_030 |
0.0781860544 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002499_070 |
0.0820771635 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 7 |
Hb_004899_070 |
0.0824376693 |
- |
- |
PREDICTED: RRP12-like protein [Jatropha curcas] |
| 8 |
Hb_000813_100 |
0.0829340033 |
- |
- |
PREDICTED: uncharacterized protein LOC105631583 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002235_320 |
0.0835001326 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001417_050 |
0.0864356425 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 11 |
Hb_005895_040 |
0.086653147 |
- |
- |
PREDICTED: tRNA threonylcarbamoyladenosine dehydratase [Jatropha curcas] |
| 12 |
Hb_000347_570 |
0.0869552086 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12b isoform X1 [Jatropha curcas] |
| 13 |
Hb_004225_130 |
0.0872337486 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105628060 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000239_070 |
0.0877541087 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001946_300 |
0.0894055334 |
- |
- |
PREDICTED: F-box protein At-B [Jatropha curcas] |
| 16 |
Hb_027073_060 |
0.0902859918 |
- |
- |
PREDICTED: digalactosyldiacylglycerol synthase 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000475_020 |
0.0904811288 |
- |
- |
PREDICTED: dehydrogenase/reductase SDR family member on chromosome X homolog isoform X2 [Jatropha curcas] |
| 18 |
Hb_000424_010 |
0.0915452942 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 15 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001771_120 |
0.0916922022 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 20 |
Hb_147737_010 |
0.0924105658 |
- |
- |
PREDICTED: arginine/serine-rich coiled-coil protein 2 isoform X2 [Jatropha curcas] |