| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
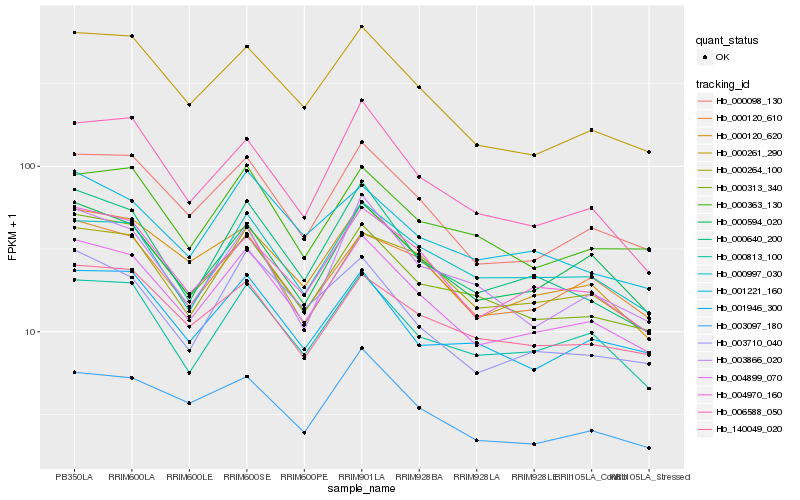
Hb_004899_070 |
0.0 |
- |
- |
PREDICTED: RRP12-like protein [Jatropha curcas] |
| 2 |
Hb_000098_130 |
0.0449218771 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2A [Jatropha curcas] |
| 3 |
Hb_000261_290 |
0.0504374519 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 4 |
Hb_000120_620 |
0.0558080191 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1a isoform X1 [Jatropha curcas] |
| 5 |
Hb_004970_160 |
0.0572247308 |
- |
- |
PREDICTED: THO complex subunit 5B-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_000640_200 |
0.063497413 |
- |
- |
hypothetical protein RCOM_1346600 [Ricinus communis] |
| 7 |
Hb_000813_100 |
0.0643366814 |
- |
- |
PREDICTED: uncharacterized protein LOC105631583 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000313_340 |
0.065991002 |
- |
- |
DNA repair protein xp-E, putative [Ricinus communis] |
| 9 |
Hb_000594_020 |
0.0678372251 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000120_610 |
0.0735541983 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 11 |
Hb_003097_180 |
0.0741647717 |
- |
- |
PREDICTED: crossover junction endonuclease MUS81 [Jatropha curcas] |
| 12 |
Hb_006588_050 |
0.0748211035 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 13 |
Hb_000264_100 |
0.0761955101 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001946_300 |
0.0767415123 |
- |
- |
PREDICTED: F-box protein At-B [Jatropha curcas] |
| 15 |
Hb_140049_020 |
0.0776356903 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 16 |
Hb_001221_160 |
0.0795339699 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 17 |
Hb_000363_130 |
0.0797964983 |
- |
- |
PREDICTED: FRIGIDA-like protein 1 [Jatropha curcas] |
| 18 |
Hb_003710_040 |
0.0805985226 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 19 |
Hb_003866_020 |
0.0806976117 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000997_030 |
0.0823493506 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |