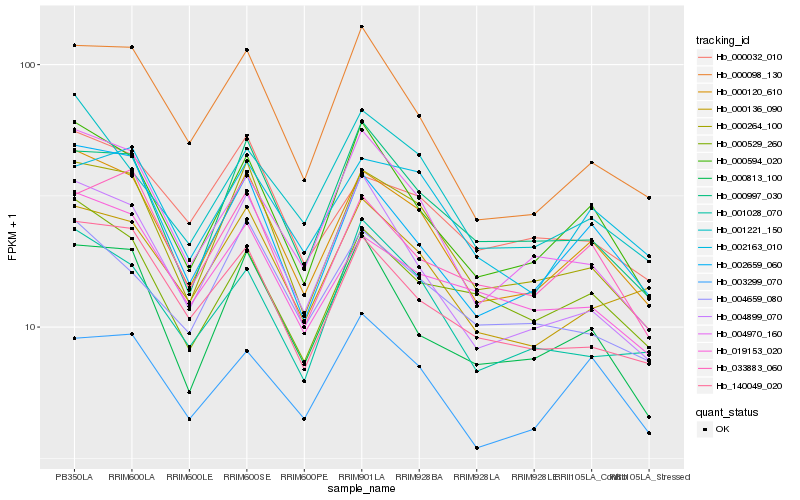
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_610 |
0.0 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 2 |
Hb_000264_100 |
0.048414288 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000594_020 |
0.0530198454 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002659_060 |
0.0595052689 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3B [Jatropha curcas] |
| 5 |
Hb_140049_020 |
0.0714540545 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 6 |
Hb_004899_070 |
0.0735541983 |
- |
- |
PREDICTED: RRP12-like protein [Jatropha curcas] |
| 7 |
Hb_004970_160 |
0.0740231226 |
- |
- |
PREDICTED: THO complex subunit 5B-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_000136_090 |
0.0740986477 |
- |
- |
programmed cell death protein, putative [Ricinus communis] |
| 9 |
Hb_001221_150 |
0.0757608441 |
- |
- |
AMP-activated protein kinase, gamma regulatory subunit, putative [Ricinus communis] |
| 10 |
Hb_000032_010 |
0.0765096964 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 11 |
Hb_019153_020 |
0.0765831545 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000813_100 |
0.0775098996 |
- |
- |
PREDICTED: uncharacterized protein LOC105631583 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000529_260 |
0.0778800798 |
- |
- |
PREDICTED: protein SGT1 homolog At5g65490 [Jatropha curcas] |
| 14 |
Hb_002163_010 |
0.0781077162 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410 [Jatropha curcas] |
| 15 |
Hb_000997_030 |
0.0783731785 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 16 |
Hb_001028_070 |
0.0785720467 |
- |
- |
PREDICTED: WD repeat-containing protein 75 isoform X2 [Jatropha curcas] |
| 17 |
Hb_033883_060 |
0.0796387512 |
- |
- |
PREDICTED: uncharacterized protein LOC105648353 [Jatropha curcas] |
| 18 |
Hb_000098_130 |
0.0796425725 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2A [Jatropha curcas] |
| 19 |
Hb_003299_070 |
0.0812470618 |
- |
- |
AMP deaminase [Theobroma cacao] |
| 20 |
Hb_004659_080 |
0.0813143663 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |