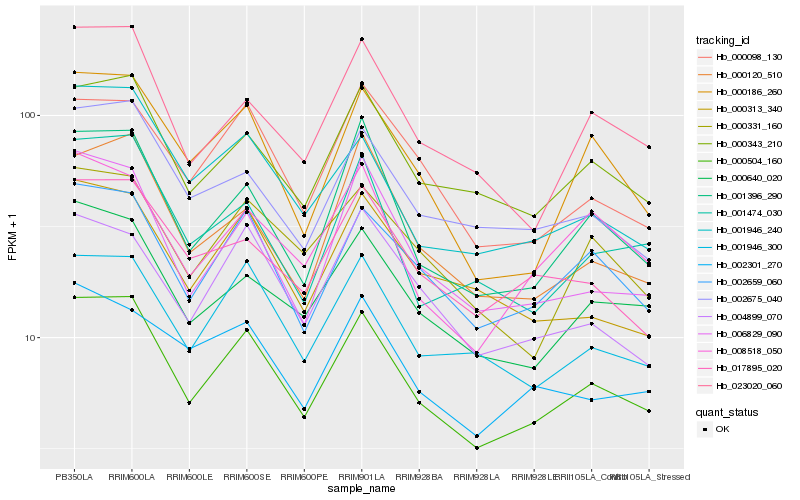
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000504_160 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s35340g [Populus trichocarpa] |
| 2 |
Hb_002659_060 |
0.0642784832 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3B [Jatropha curcas] |
| 3 |
Hb_001396_290 |
0.0700982384 |
- |
- |
hypothetical protein B456_005G166600 [Gossypium raimondii] |
| 4 |
Hb_000343_210 |
0.0705505556 |
- |
- |
ran-binding protein, putative [Ricinus communis] |
| 5 |
Hb_001946_300 |
0.075013453 |
- |
- |
PREDICTED: F-box protein At-B [Jatropha curcas] |
| 6 |
Hb_000640_020 |
0.0752995338 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 7 |
Hb_006829_090 |
0.0783980884 |
- |
- |
PREDICTED: protein MOS2 [Jatropha curcas] |
| 8 |
Hb_001946_240 |
0.0794685713 |
- |
- |
PREDICTED: chaperone protein dnaJ 49-like [Jatropha curcas] |
| 9 |
Hb_023020_060 |
0.0796213814 |
- |
- |
ubiquitin-conjugating enzyme e2S, putative [Ricinus communis] |
| 10 |
Hb_000186_260 |
0.079699811 |
transcription factor |
TF Family: PHD |
PREDICTED: protein polybromo-1-like [Jatropha curcas] |
| 11 |
Hb_002675_040 |
0.0830978202 |
- |
- |
ran-binding protein, putative [Ricinus communis] |
| 12 |
Hb_000098_130 |
0.0855782957 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2A [Jatropha curcas] |
| 13 |
Hb_000120_510 |
0.0859872846 |
- |
- |
PREDICTED: RNA polymerase I-specific transcription initiation factor RRN3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001474_030 |
0.0860061404 |
- |
- |
PREDICTED: uncharacterized protein LOC105650916 [Jatropha curcas] |
| 15 |
Hb_000331_160 |
0.0869990186 |
- |
- |
adenylate kinase 1 chloroplast, putative [Ricinus communis] |
| 16 |
Hb_008518_050 |
0.0885810006 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 17 |
Hb_000313_340 |
0.0910448838 |
- |
- |
DNA repair protein xp-E, putative [Ricinus communis] |
| 18 |
Hb_017895_020 |
0.091186521 |
- |
- |
Vacuolar protein sorting protein, putative [Ricinus communis] |
| 19 |
Hb_002301_270 |
0.0914694371 |
- |
- |
PREDICTED: glyoxysomal processing protease, glyoxysomal isoform X1 [Jatropha curcas] |
| 20 |
Hb_004899_070 |
0.0921664608 |
- |
- |
PREDICTED: RRP12-like protein [Jatropha curcas] |