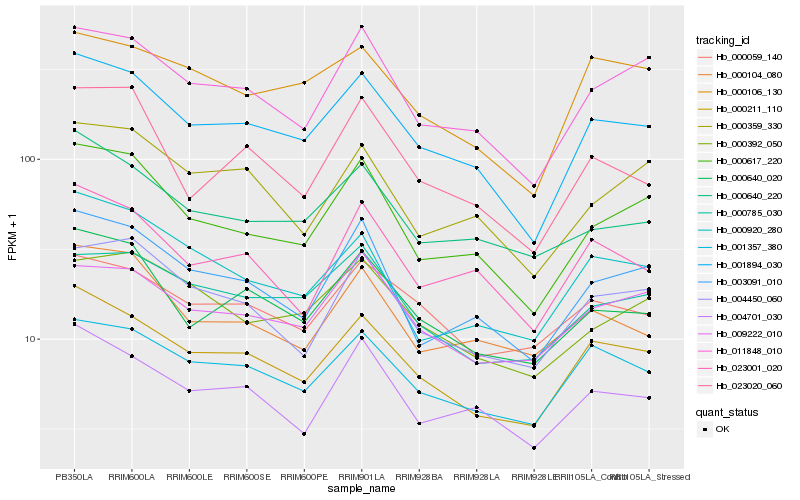
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001894_030 |
0.0 |
- |
- |
Nucleic acid-binding, OB-fold-like protein [Theobroma cacao] |
| 2 |
Hb_000211_110 |
0.0561668307 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 35 kDa protein isoform X1 [Jatropha curcas] |
| 3 |
Hb_000617_220 |
0.0682228476 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 4 |
Hb_000640_020 |
0.0715026641 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 5 |
Hb_001357_380 |
0.0733525537 |
- |
- |
PREDICTED: probable tetraacyldisaccharide 4'-kinase, mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_011848_010 |
0.0767786753 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 7 |
Hb_004701_030 |
0.0796153086 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 8 |
Hb_003091_010 |
0.0797600845 |
- |
- |
PREDICTED: uncharacterized protein LOC105131887 [Populus euphratica] |
| 9 |
Hb_023001_020 |
0.0828029166 |
- |
- |
PREDICTED: mitochondrial succinate-fumarate transporter 1 [Jatropha curcas] |
| 10 |
Hb_000640_220 |
0.085788779 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-4 [Jatropha curcas] |
| 11 |
Hb_000106_130 |
0.0889128621 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_023020_060 |
0.0893993053 |
- |
- |
ubiquitin-conjugating enzyme e2S, putative [Ricinus communis] |
| 13 |
Hb_000785_030 |
0.0896055954 |
- |
- |
hypothetical protein RCOM_0808030 [Ricinus communis] |
| 14 |
Hb_004450_060 |
0.0904658027 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000104_080 |
0.090848042 |
- |
- |
PREDICTED: WD repeat domain-containing protein 83 [Jatropha curcas] |
| 16 |
Hb_009222_010 |
0.0928125484 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 17 |
Hb_000059_140 |
0.093487849 |
- |
- |
rnase h, putative [Ricinus communis] |
| 18 |
Hb_000359_330 |
0.0937098598 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000392_050 |
0.0939598067 |
- |
- |
PREDICTED: uncharacterized protein LOC105640540 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000920_280 |
0.0944654286 |
- |
- |
Protein SENSITIVITY TO RED LIGHT REDUCED, putative [Ricinus communis] |