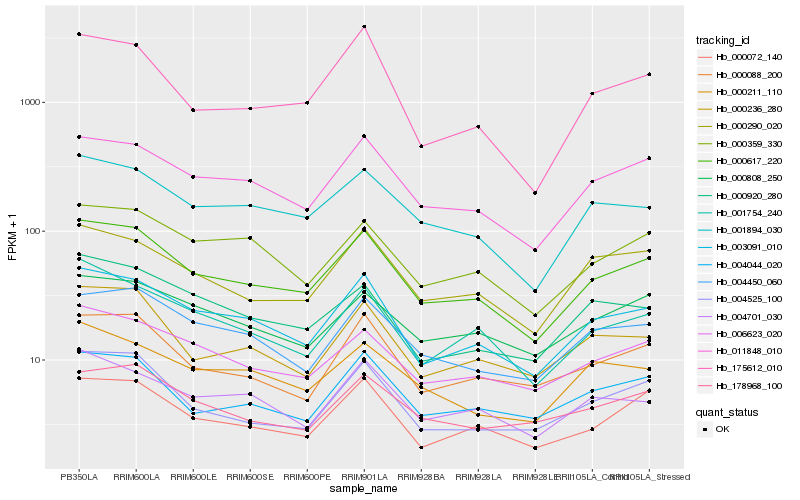
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000617_220 |
0.0 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 2 |
Hb_003091_010 |
0.0517206629 |
- |
- |
PREDICTED: uncharacterized protein LOC105131887 [Populus euphratica] |
| 3 |
Hb_011848_010 |
0.0574608636 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 4 |
Hb_001894_030 |
0.0682228476 |
- |
- |
Nucleic acid-binding, OB-fold-like protein [Theobroma cacao] |
| 5 |
Hb_006623_020 |
0.0722149019 |
- |
- |
PREDICTED: protein LURP-one-related 7 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000072_140 |
0.0740438976 |
- |
- |
- |
| 7 |
Hb_000290_020 |
0.0765183589 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit K-like isoform X2 [Populus euphratica] |
| 8 |
Hb_004525_100 |
0.0770707189 |
- |
- |
hypothetical protein JCGZ_18418 [Jatropha curcas] |
| 9 |
Hb_000088_200 |
0.0797017696 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_000359_330 |
0.0799406464 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001754_240 |
0.0799949331 |
- |
- |
PREDICTED: mitochondrial thiamine pyrophosphate carrier isoform X1 [Jatropha curcas] |
| 12 |
Hb_004450_060 |
0.0802194938 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000236_280 |
0.0822928301 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 22 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_004044_020 |
0.0837493094 |
- |
- |
PREDICTED: nudix hydrolase 19, chloroplastic [Jatropha curcas] |
| 15 |
Hb_004701_030 |
0.0864101471 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 16 |
Hb_000808_250 |
0.0882213154 |
- |
- |
PREDICTED: uncharacterized protein LOC105647419 [Jatropha curcas] |
| 17 |
Hb_000211_110 |
0.0901353168 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 35 kDa protein isoform X1 [Jatropha curcas] |
| 18 |
Hb_000920_280 |
0.0912263475 |
- |
- |
Protein SENSITIVITY TO RED LIGHT REDUCED, putative [Ricinus communis] |
| 19 |
Hb_175612_010 |
0.0921287327 |
transcription factor |
TF Family: MBF1 |
orf [Ricinus communis] |
| 20 |
Hb_178968_100 |
0.0921990115 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase isoform X1 [Populus euphratica] |