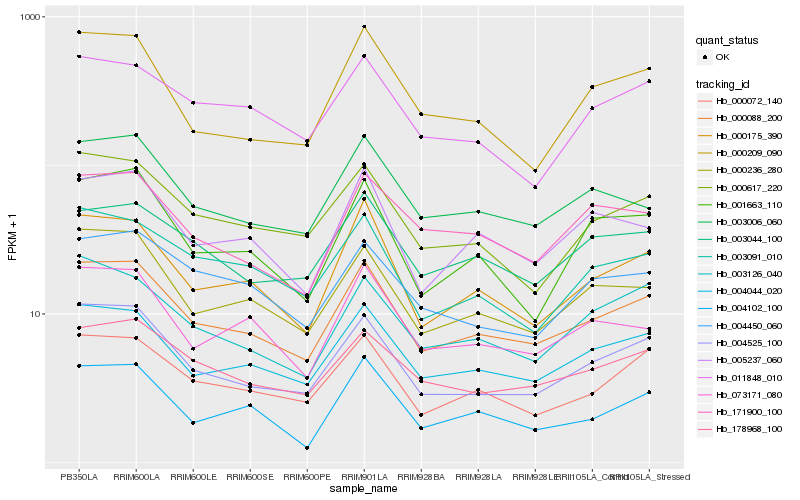
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000088_200 |
0.0 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_004044_020 |
0.049691097 |
- |
- |
PREDICTED: nudix hydrolase 19, chloroplastic [Jatropha curcas] |
| 3 |
Hb_004525_100 |
0.058014754 |
- |
- |
hypothetical protein JCGZ_18418 [Jatropha curcas] |
| 4 |
Hb_000236_280 |
0.0682004634 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 22 homolog isoform X1 [Jatropha curcas] |
| 5 |
Hb_000072_140 |
0.0686052469 |
- |
- |
- |
| 6 |
Hb_178968_100 |
0.0705512141 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase isoform X1 [Populus euphratica] |
| 7 |
Hb_000175_390 |
0.0711683508 |
- |
- |
DNA-directed RNA polymerase III subunit F, putative [Ricinus communis] |
| 8 |
Hb_003006_060 |
0.0773015833 |
- |
- |
PREDICTED: pre-mRNA-splicing factor syf2 [Jatropha curcas] |
| 9 |
Hb_005237_060 |
0.079487264 |
transcription factor |
TF Family: ERF |
ERF/AP2 domain-containing transcription factor [Manihot esculenta] |
| 10 |
Hb_000617_220 |
0.0797017696 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 11 |
Hb_003126_040 |
0.0841135482 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_073171_080 |
0.0842411108 |
- |
- |
PREDICTED: MMS19 nucleotide excision repair protein homolog isoform X1 [Jatropha curcas] |
| 13 |
Hb_003044_100 |
0.084869864 |
- |
- |
hypothetical protein F383_08446 [Gossypium arboreum] |
| 14 |
Hb_003091_010 |
0.0856614903 |
- |
- |
PREDICTED: uncharacterized protein LOC105131887 [Populus euphratica] |
| 15 |
Hb_000209_090 |
0.0896481125 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 16 |
Hb_001663_110 |
0.0899788907 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_004102_100 |
0.0914922619 |
- |
- |
PREDICTED: periodic tryptophan protein 2 homolog [Jatropha curcas] |
| 18 |
Hb_171900_100 |
0.0916270078 |
- |
- |
PREDICTED: putative uncharacterized protein DDB_G0270496 [Jatropha curcas] |
| 19 |
Hb_004450_060 |
0.0918579809 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X2 [Jatropha curcas] |
| 20 |
Hb_011848_010 |
0.0922853311 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |