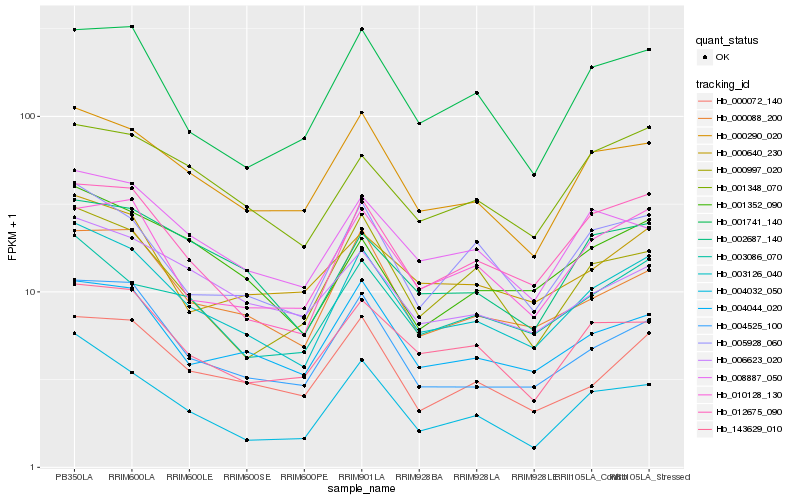
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003126_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000290_020 |
0.0780530248 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit K-like isoform X2 [Populus euphratica] |
| 3 |
Hb_012675_090 |
0.0788582513 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 4-like isoform X1 [Citrus sinensis] |
| 4 |
Hb_004525_100 |
0.079396876 |
- |
- |
hypothetical protein JCGZ_18418 [Jatropha curcas] |
| 5 |
Hb_004044_020 |
0.0836109836 |
- |
- |
PREDICTED: nudix hydrolase 19, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003086_070 |
0.0837421664 |
- |
- |
PREDICTED: iron-sulfur cluster co-chaperone protein HscB, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000088_200 |
0.0841135482 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_005928_060 |
0.0890129946 |
- |
- |
DNA-directed RNA polymerase III 25 kD polypeptide, putative [Ricinus communis] |
| 9 |
Hb_143629_010 |
0.0891455422 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 10 |
Hb_000072_140 |
0.0938051329 |
- |
- |
- |
| 11 |
Hb_004032_050 |
0.0941449524 |
- |
- |
hypothetical protein CISIN_1g014592mg [Citrus sinensis] |
| 12 |
Hb_008887_050 |
0.0946556084 |
- |
- |
PREDICTED: DNA-binding protein BIN4 [Jatropha curcas] |
| 13 |
Hb_010128_130 |
0.0951345546 |
- |
- |
PREDICTED: G patch domain-containing protein 8 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001352_090 |
0.0981972255 |
- |
- |
PREDICTED: UPF0161 protein At3g09310 [Jatropha curcas] |
| 15 |
Hb_000997_020 |
0.0982119009 |
- |
- |
PREDICTED: uncharacterized protein LOC105629694 [Jatropha curcas] |
| 16 |
Hb_002687_140 |
0.0985913039 |
- |
- |
hypothetical protein JCGZ_24463 [Jatropha curcas] |
| 17 |
Hb_000640_230 |
0.0986923489 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_006623_020 |
0.0987523154 |
- |
- |
PREDICTED: protein LURP-one-related 7 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001741_140 |
0.0989197953 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 20 |
Hb_001348_070 |
0.0993482019 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |