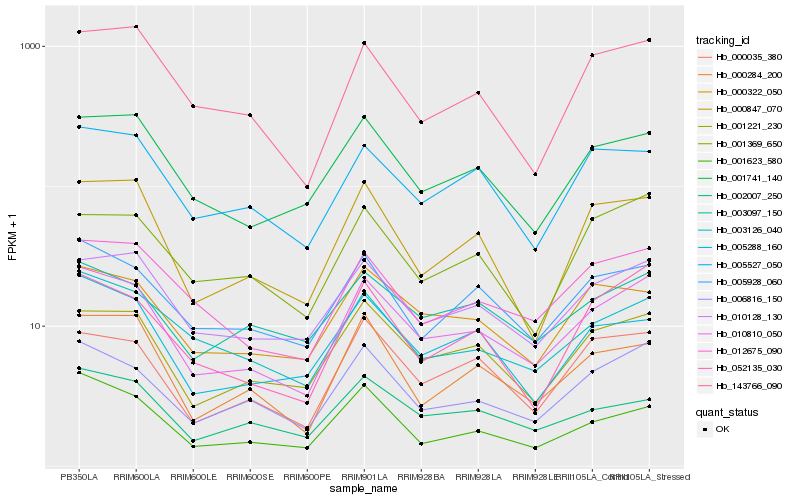
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010810_050 |
0.0 |
- |
- |
hypothetical protein [Cleome spinosa] |
| 2 |
Hb_006816_150 |
0.0789762131 |
- |
- |
PREDICTED: lon protease homolog 1, mitochondrial [Jatropha curcas] |
| 3 |
Hb_001221_230 |
0.0867706466 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_010128_130 |
0.0902949625 |
- |
- |
PREDICTED: G patch domain-containing protein 8 isoform X1 [Jatropha curcas] |
| 5 |
Hb_012675_090 |
0.0965347675 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 4-like isoform X1 [Citrus sinensis] |
| 6 |
Hb_052135_030 |
0.097869781 |
- |
- |
PREDICTED: huntingtin-interacting protein K [Jatropha curcas] |
| 7 |
Hb_005928_060 |
0.0978831715 |
- |
- |
DNA-directed RNA polymerase III 25 kD polypeptide, putative [Ricinus communis] |
| 8 |
Hb_000035_380 |
0.0983732492 |
- |
- |
- |
| 9 |
Hb_001741_140 |
0.0990239661 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 10 |
Hb_003126_040 |
0.1005580521 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000847_070 |
0.1042528249 |
- |
- |
PREDICTED: uncharacterized protein LOC105125699 [Populus euphratica] |
| 12 |
Hb_000322_050 |
0.1054290155 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 13 |
Hb_143766_090 |
0.1057870591 |
- |
- |
60S ribosomal protein L31B [Hevea brasiliensis] |
| 14 |
Hb_005527_050 |
0.1078636383 |
- |
- |
PREDICTED: uncharacterized protein LOC105643024 [Jatropha curcas] |
| 15 |
Hb_005288_160 |
0.1083445896 |
- |
- |
PREDICTED: uncharacterized protein LOC105647746 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003097_150 |
0.109065536 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 24 [Jatropha curcas] |
| 17 |
Hb_001623_580 |
0.1090723913 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase family member SUVH9-like [Jatropha curcas] |
| 18 |
Hb_002007_250 |
0.1092079021 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000284_200 |
0.1103998281 |
- |
- |
- |
| 20 |
Hb_001369_650 |
0.1107857431 |
- |
- |
PREDICTED: chloride conductance regulatory protein ICln [Jatropha curcas] |