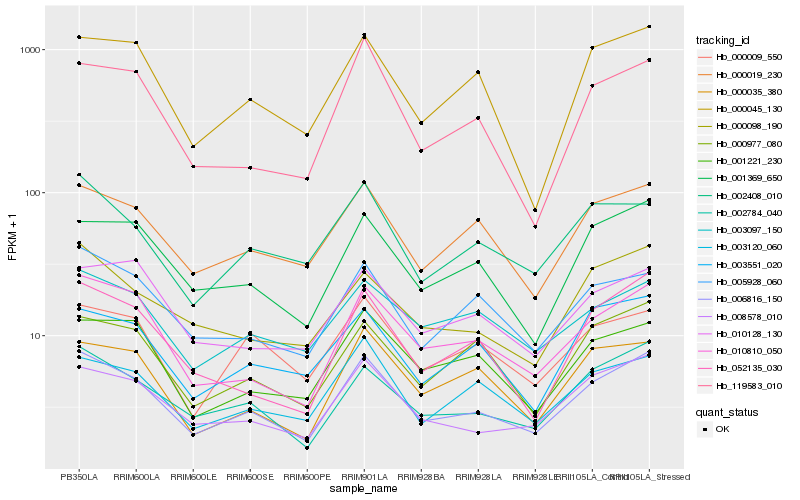
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006816_150 |
0.0 |
- |
- |
PREDICTED: lon protease homolog 1, mitochondrial [Jatropha curcas] |
| 2 |
Hb_002784_040 |
0.0735530745 |
- |
- |
peptidyl-prolyl cis-trans isomerase h, ppih, putative [Ricinus communis] |
| 3 |
Hb_010810_050 |
0.0789762131 |
- |
- |
hypothetical protein [Cleome spinosa] |
| 4 |
Hb_008578_010 |
0.0792009514 |
- |
- |
30S ribosomal protein S19, putative [Ricinus communis] |
| 5 |
Hb_000019_230 |
0.0900808613 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit Rieske-4, mitochondrial-like [Jatropha curcas] |
| 6 |
Hb_001369_650 |
0.0932690246 |
- |
- |
PREDICTED: chloride conductance regulatory protein ICln [Jatropha curcas] |
| 7 |
Hb_003120_060 |
0.1007818489 |
- |
- |
PREDICTED: prostaglandin E synthase 2 [Jatropha curcas] |
| 8 |
Hb_005928_060 |
0.1016197942 |
- |
- |
DNA-directed RNA polymerase III 25 kD polypeptide, putative [Ricinus communis] |
| 9 |
Hb_000977_080 |
0.1019431346 |
- |
- |
type II inositol 5-phosphatase, putative [Ricinus communis] |
| 10 |
Hb_052135_030 |
0.1021472346 |
- |
- |
PREDICTED: huntingtin-interacting protein K [Jatropha curcas] |
| 11 |
Hb_000098_190 |
0.1022687272 |
- |
- |
PREDICTED: uncharacterized protein LOC105633343 [Jatropha curcas] |
| 12 |
Hb_003097_150 |
0.1044033349 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 24 [Jatropha curcas] |
| 13 |
Hb_003551_020 |
0.1047832763 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 3-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001221_230 |
0.1060223803 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000045_130 |
0.1069783912 |
- |
- |
60S acidic ribosomal protein P0A [Hevea brasiliensis] |
| 16 |
Hb_002408_010 |
0.1070144617 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 17 |
Hb_000035_380 |
0.1086709209 |
- |
- |
- |
| 18 |
Hb_000009_550 |
0.1105807235 |
- |
- |
hypothetical protein RCOM_0603640 [Ricinus communis] |
| 19 |
Hb_119583_010 |
0.112016461 |
- |
- |
60S ribosomal protein L12A [Hevea brasiliensis] |
| 20 |
Hb_010128_130 |
0.1130440032 |
- |
- |
PREDICTED: G patch domain-containing protein 8 isoform X1 [Jatropha curcas] |