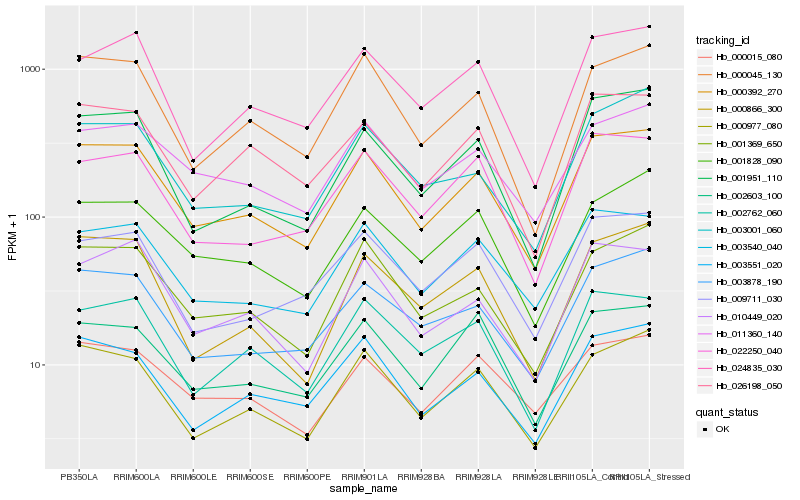
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000392_270 |
0.0 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 2 |
Hb_000977_080 |
0.0594319176 |
- |
- |
type II inositol 5-phosphatase, putative [Ricinus communis] |
| 3 |
Hb_001369_650 |
0.0635900919 |
- |
- |
PREDICTED: chloride conductance regulatory protein ICln [Jatropha curcas] |
| 4 |
Hb_003540_040 |
0.0637444708 |
- |
- |
hypothetical protein JCGZ_20979 [Jatropha curcas] |
| 5 |
Hb_003551_020 |
0.0648369178 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 3-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_003878_190 |
0.064968586 |
- |
- |
hypothetical protein POPTR_0011s11040g [Populus trichocarpa] |
| 7 |
Hb_011360_140 |
0.070913732 |
- |
- |
40S ribosomal protein S19, putative [Ricinus communis] |
| 8 |
Hb_001951_110 |
0.0711222192 |
- |
- |
PREDICTED: DNA-binding protein DDB_G0278111 [Jatropha curcas] |
| 9 |
Hb_026198_050 |
0.0711374501 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.3 [Jatropha curcas] |
| 10 |
Hb_010449_020 |
0.071315042 |
- |
- |
PREDICTED: rRNA-processing protein UTP23 homolog [Jatropha curcas] |
| 11 |
Hb_000045_130 |
0.0723793974 |
- |
- |
60S acidic ribosomal protein P0A [Hevea brasiliensis] |
| 12 |
Hb_001828_090 |
0.073487004 |
- |
- |
prohibitin, putative [Ricinus communis] |
| 13 |
Hb_002762_060 |
0.0759822435 |
- |
- |
PREDICTED: diphthine methyltransferase homolog [Jatropha curcas] |
| 14 |
Hb_022250_040 |
0.0765054375 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 2, mitochondrial [Jatropha curcas] |
| 15 |
Hb_024835_030 |
0.0765490099 |
- |
- |
ubiquitin-conjugating enzyme E2, putative [Ricinus communis] |
| 16 |
Hb_000866_300 |
0.0788371145 |
- |
- |
hypothetical protein CICLE_v10023145mg [Citrus clementina] |
| 17 |
Hb_000015_080 |
0.0788724053 |
- |
- |
PREDICTED: uncharacterized protein LOC105647918 isoform X1 [Jatropha curcas] |
| 18 |
Hb_009711_030 |
0.0799704871 |
- |
- |
PREDICTED: uncharacterized protein LOC105647792 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003001_060 |
0.0800529431 |
- |
- |
wound-responsive family protein [Populus trichocarpa] |
| 20 |
Hb_002603_100 |
0.0803463263 |
- |
- |
ATATH9, putative [Ricinus communis] |