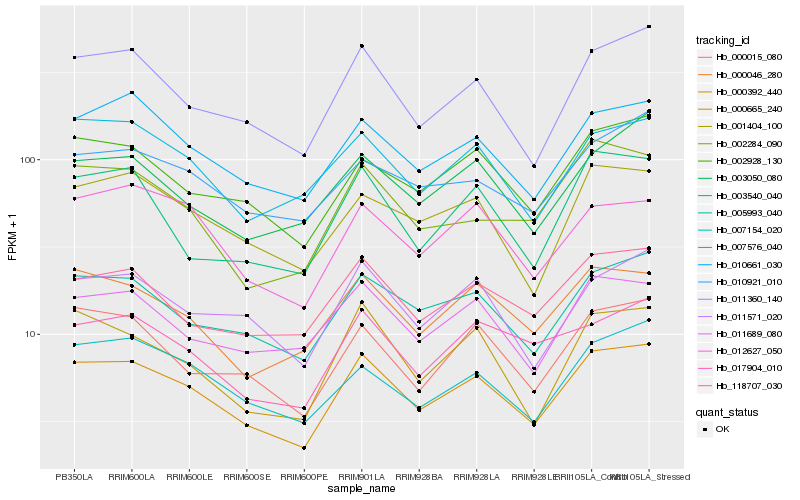
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000392_440 |
0.0 |
- |
- |
PREDICTED: nuclear ribonuclease Z isoform X1 [Jatropha curcas] |
| 2 |
Hb_011360_140 |
0.060978694 |
- |
- |
40S ribosomal protein S19, putative [Ricinus communis] |
| 3 |
Hb_005993_040 |
0.0639489211 |
- |
- |
PREDICTED: tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit wdr4 [Jatropha curcas] |
| 4 |
Hb_010661_030 |
0.070582268 |
- |
- |
hypothetical protein PHAVU_009G146200g [Phaseolus vulgaris] |
| 5 |
Hb_002928_130 |
0.0724840981 |
- |
- |
PREDICTED: small nuclear ribonucleoprotein Sm D3 [Jatropha curcas] |
| 6 |
Hb_000015_080 |
0.0747787511 |
- |
- |
PREDICTED: uncharacterized protein LOC105647918 isoform X1 [Jatropha curcas] |
| 7 |
Hb_011571_020 |
0.0750614121 |
- |
- |
PREDICTED: metal tolerance protein C4 [Jatropha curcas] |
| 8 |
Hb_000046_280 |
0.0756454233 |
- |
- |
PREDICTED: uncharacterized protein LOC105631831 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001404_100 |
0.0782036604 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit K [Gossypium raimondii] |
| 10 |
Hb_000665_240 |
0.0784741166 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |
| 11 |
Hb_007576_040 |
0.079241453 |
- |
- |
PREDICTED: uncharacterized protein LOC105169221 [Sesamum indicum] |
| 12 |
Hb_118707_030 |
0.0796594142 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 19a-like isoform X1 [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_011689_080 |
0.07968508 |
- |
- |
PREDICTED: uncharacterized protein LOC105631867 [Jatropha curcas] |
| 14 |
Hb_007154_020 |
0.0811614273 |
- |
- |
- |
| 15 |
Hb_012627_050 |
0.0857996369 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_003540_040 |
0.0862888925 |
- |
- |
hypothetical protein JCGZ_20979 [Jatropha curcas] |
| 17 |
Hb_017904_010 |
0.0873853181 |
- |
- |
PREDICTED: DNA repair protein recA homolog 2, mitochondrial [Jatropha curcas] |
| 18 |
Hb_010921_010 |
0.0887584345 |
- |
- |
Small nuclear ribonucleoprotein family protein [Theobroma cacao] |
| 19 |
Hb_002284_090 |
0.0899145664 |
- |
- |
PREDICTED: outer envelope pore protein 21B, chloroplastic [Jatropha curcas] |
| 20 |
Hb_003050_080 |
0.0900823038 |
- |
- |
40S ribosomal protein S14, putative [Ricinus communis] |