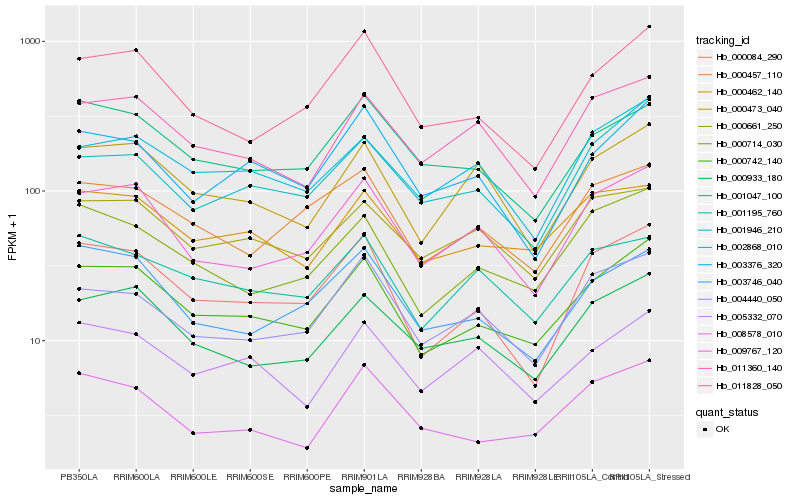
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000742_140 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000714_030 |
0.0751189461 |
- |
- |
Nucleosome assembly [Gossypium arboreum] |
| 3 |
Hb_000084_290 |
0.0772672786 |
- |
- |
mitochondrial 39S ribosomal protein L53 [Hevea brasiliensis] |
| 4 |
Hb_000933_180 |
0.079048151 |
- |
- |
PREDICTED: chorismate mutase 3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_009767_120 |
0.0794329049 |
- |
- |
PREDICTED: probable prefoldin subunit 2 [Jatropha curcas] |
| 6 |
Hb_011828_050 |
0.0822497311 |
- |
- |
40S ribosomal protein S23, putative [Ricinus communis] |
| 7 |
Hb_000462_140 |
0.0868615589 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 28 [Jatropha curcas] |
| 8 |
Hb_000473_040 |
0.0894695767 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: RNA polymerase II transcriptional coactivator KIWI-like [Jatropha curcas] |
| 9 |
Hb_000457_110 |
0.0914140144 |
- |
- |
Mitochondrial ribosomal protein L37 isoform 1 [Theobroma cacao] |
| 10 |
Hb_003746_040 |
0.0918284215 |
- |
- |
PREDICTED: protein N-terminal glutamine amidohydrolase [Jatropha curcas] |
| 11 |
Hb_002868_010 |
0.0928839096 |
- |
- |
60S ribosomal protein L30, putative [Ricinus communis] |
| 12 |
Hb_011360_140 |
0.0934196677 |
- |
- |
40S ribosomal protein S19, putative [Ricinus communis] |
| 13 |
Hb_004440_050 |
0.0949942512 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 10 [Cucumis melo] |
| 14 |
Hb_008578_010 |
0.095196106 |
- |
- |
30S ribosomal protein S19, putative [Ricinus communis] |
| 15 |
Hb_001047_100 |
0.0952702937 |
- |
- |
40S ribosomal protein S18, putative [Ricinus communis] |
| 16 |
Hb_003376_320 |
0.0955334728 |
- |
- |
60S ribosomal protein L12A [Hevea brasiliensis] |
| 17 |
Hb_001946_210 |
0.0965722197 |
- |
- |
ribosomal protein l7ae, putative [Ricinus communis] |
| 18 |
Hb_001195_760 |
0.0966941632 |
- |
- |
PREDICTED: 30S ribosomal protein S9, chloroplastic-like [Populus euphratica] |
| 19 |
Hb_000661_250 |
0.0975282478 |
- |
- |
PREDICTED: RNA-binding protein 8A [Jatropha curcas] |
| 20 |
Hb_005332_070 |
0.0978043685 |
- |
- |
PREDICTED: uncharacterized protein LOC105632487 [Jatropha curcas] |