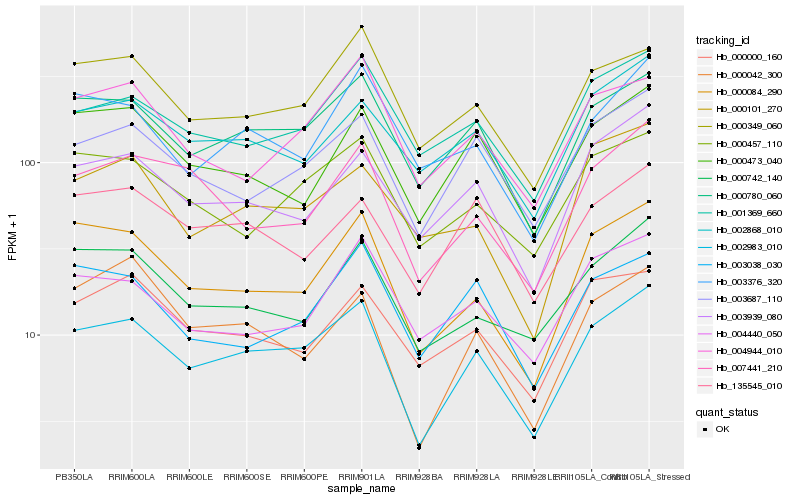
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002983_010 |
0.0 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 2 |
Hb_000780_060 |
0.0729357805 |
- |
- |
hypothetical protein CICLE_v10017178mg [Citrus clementina] |
| 3 |
Hb_000084_290 |
0.0906531438 |
- |
- |
mitochondrial 39S ribosomal protein L53 [Hevea brasiliensis] |
| 4 |
Hb_003687_110 |
0.0943972304 |
- |
- |
PREDICTED: uncharacterized protein At4g14342 isoform X1 [Malus domestica] |
| 5 |
Hb_000349_060 |
0.1010880607 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 6 |
Hb_003939_080 |
0.1031810948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001369_660 |
0.1070968482 |
- |
- |
hypothetical protein B456_001G174200, partial [Gossypium raimondii] |
| 8 |
Hb_000457_110 |
0.107645784 |
- |
- |
Mitochondrial ribosomal protein L37 isoform 1 [Theobroma cacao] |
| 9 |
Hb_000742_140 |
0.1100693868 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007441_210 |
0.1105522395 |
- |
- |
hypothetical protein PRUPE_ppa013350mg [Prunus persica] |
| 11 |
Hb_002868_010 |
0.1105940771 |
- |
- |
60S ribosomal protein L30, putative [Ricinus communis] |
| 12 |
Hb_000473_040 |
0.1123610343 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: RNA polymerase II transcriptional coactivator KIWI-like [Jatropha curcas] |
| 13 |
Hb_003376_320 |
0.1130890036 |
- |
- |
60S ribosomal protein L12A [Hevea brasiliensis] |
| 14 |
Hb_135545_010 |
0.1140520738 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 15 |
Hb_004944_010 |
0.1163670414 |
- |
- |
small nuclear ribonucleoprotein E [Hevea brasiliensis] |
| 16 |
Hb_000000_160 |
0.1164535063 |
- |
- |
PREDICTED: membrin-11 [Jatropha curcas] |
| 17 |
Hb_004440_050 |
0.1193696001 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 10 [Cucumis melo] |
| 18 |
Hb_003038_030 |
0.11989554 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 10 [Cucumis melo] |
| 19 |
Hb_000042_300 |
0.1213060867 |
- |
- |
- |
| 20 |
Hb_000101_270 |
0.121433491 |
- |
- |
pleckstrin homology domain-containing family protein [Populus trichocarpa] |