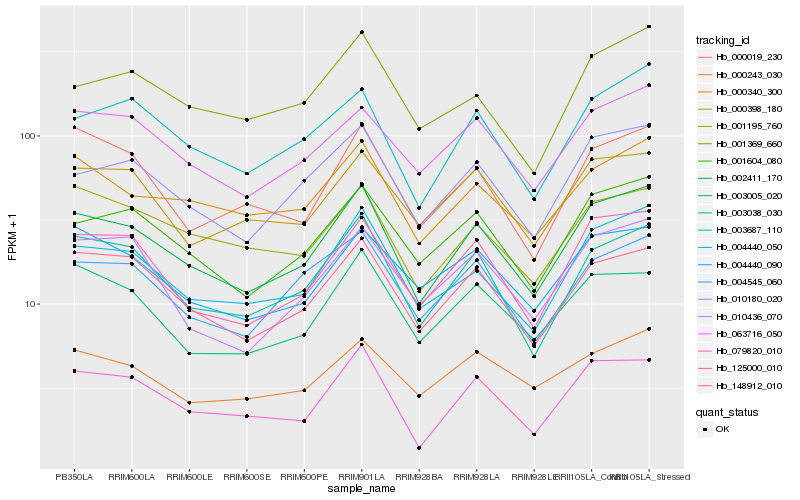
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002411_170 |
0.0 |
- |
- |
PREDICTED: cilia- and flagella-associated protein 20 [Jatropha curcas] |
| 2 |
Hb_004440_050 |
0.05745372 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 10 [Cucumis melo] |
| 3 |
Hb_003038_030 |
0.0613135801 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 10 [Cucumis melo] |
| 4 |
Hb_125000_010 |
0.0622346625 |
- |
- |
PREDICTED: protein SCO1 homolog 2, mitochondrial [Jatropha curcas] |
| 5 |
Hb_148912_010 |
0.0633045795 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_010180_020 |
0.0637832803 |
- |
- |
hypothetical protein glysoja_030394 [Glycine soja] |
| 7 |
Hb_001604_080 |
0.0638345181 |
- |
- |
PREDICTED: uncharacterized protein LOC105638437 [Jatropha curcas] |
| 8 |
Hb_079820_010 |
0.069762105 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 1 [Jatropha curcas] |
| 9 |
Hb_063716_050 |
0.0739378881 |
- |
- |
PREDICTED: uncharacterized protein LOC105634866 [Jatropha curcas] |
| 10 |
Hb_000340_300 |
0.0739598528 |
- |
- |
methyl-CpG-binding domain-containing family protein [Populus trichocarpa] |
| 11 |
Hb_003687_110 |
0.074599331 |
- |
- |
PREDICTED: uncharacterized protein At4g14342 isoform X1 [Malus domestica] |
| 12 |
Hb_004545_060 |
0.0768726148 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 6B [Jatropha curcas] |
| 13 |
Hb_003005_020 |
0.0794025611 |
- |
- |
PREDICTED: DNA repair protein XRCC3 homolog [Jatropha curcas] |
| 14 |
Hb_004440_090 |
0.0833798042 |
- |
- |
PREDICTED: phosphopantetheine adenylyltransferase [Jatropha curcas] |
| 15 |
Hb_000398_180 |
0.0844289331 |
- |
- |
hypothetical protein PRUPE_ppa009665mg [Prunus persica] |
| 16 |
Hb_000243_030 |
0.0846434756 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 17 |
Hb_010436_070 |
0.0850097878 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 4-like [Nicotiana tomentosiformis] |
| 18 |
Hb_001195_760 |
0.0854736975 |
- |
- |
PREDICTED: 30S ribosomal protein S9, chloroplastic-like [Populus euphratica] |
| 19 |
Hb_001369_660 |
0.0855935864 |
- |
- |
hypothetical protein B456_001G174200, partial [Gossypium raimondii] |
| 20 |
Hb_000019_230 |
0.0866341486 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit Rieske-4, mitochondrial-like [Jatropha curcas] |