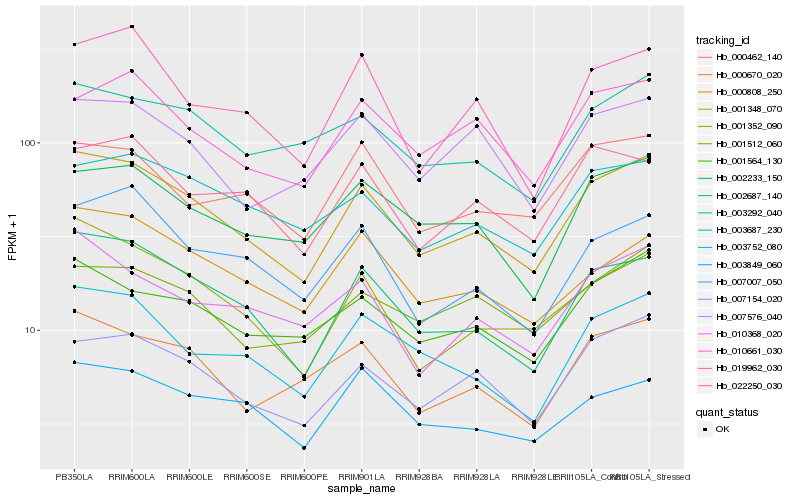
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001348_070 |
0.0 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 2 |
Hb_002687_140 |
0.0562343074 |
- |
- |
hypothetical protein JCGZ_24463 [Jatropha curcas] |
| 3 |
Hb_007154_020 |
0.0692127385 |
- |
- |
- |
| 4 |
Hb_003752_080 |
0.0757472168 |
- |
- |
PREDICTED: ubiquitin-like-conjugating enzyme ATG10 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001564_130 |
0.0766432044 |
- |
- |
PREDICTED: single-stranded DNA-binding protein, mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_000670_020 |
0.0797553262 |
- |
- |
PREDICTED: chromatin modification-related protein EAF3-like [Jatropha curcas] |
| 7 |
Hb_002233_150 |
0.0800641608 |
- |
- |
PREDICTED: uncharacterized protein LOC105633093 [Jatropha curcas] |
| 8 |
Hb_001352_090 |
0.0817076663 |
- |
- |
PREDICTED: UPF0161 protein At3g09310 [Jatropha curcas] |
| 9 |
Hb_000808_250 |
0.0822488531 |
- |
- |
PREDICTED: uncharacterized protein LOC105647419 [Jatropha curcas] |
| 10 |
Hb_003292_040 |
0.085582387 |
transcription factor |
TF Family: MBF1 |
uncharacterized protein LOC100500420 [Glycine max] |
| 11 |
Hb_003687_230 |
0.0867847565 |
- |
- |
PREDICTED: actin-depolymerizing factor-like [Jatropha curcas] |
| 12 |
Hb_019962_030 |
0.0871475832 |
- |
- |
PREDICTED: uncharacterized protein LOC105650622 [Jatropha curcas] |
| 13 |
Hb_010661_030 |
0.0873622015 |
- |
- |
hypothetical protein PHAVU_009G146200g [Phaseolus vulgaris] |
| 14 |
Hb_010368_020 |
0.0876602702 |
- |
- |
PREDICTED: uncharacterized protein LOC105630301 [Jatropha curcas] |
| 15 |
Hb_007007_050 |
0.0877042551 |
- |
- |
PREDICTED: ubiquitin-related modifier 1 homolog 2 [Malus domestica] |
| 16 |
Hb_003849_060 |
0.08818032 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 17 |
Hb_022250_030 |
0.088731763 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000462_140 |
0.0898114821 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 28 [Jatropha curcas] |
| 19 |
Hb_001512_060 |
0.0918645722 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g35850, mitochondrial [Jatropha curcas] |
| 20 |
Hb_007576_040 |
0.0924170175 |
- |
- |
PREDICTED: uncharacterized protein LOC105169221 [Sesamum indicum] |