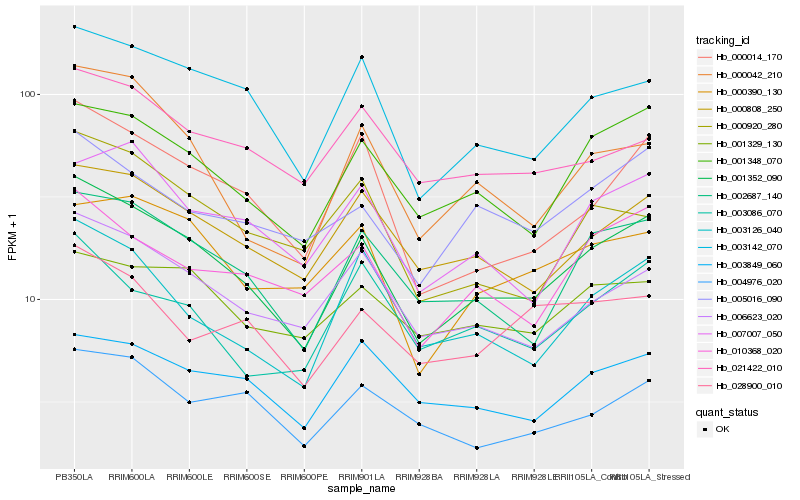
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001352_090 |
0.0 |
- |
- |
PREDICTED: UPF0161 protein At3g09310 [Jatropha curcas] |
| 2 |
Hb_002687_140 |
0.0800430672 |
- |
- |
hypothetical protein JCGZ_24463 [Jatropha curcas] |
| 3 |
Hb_001348_070 |
0.0817076663 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 4 |
Hb_003142_070 |
0.0841341549 |
- |
- |
hypothetical protein F383_14657 [Gossypium arboreum] |
| 5 |
Hb_006623_020 |
0.0912196662 |
- |
- |
PREDICTED: protein LURP-one-related 7 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000808_250 |
0.0917523254 |
- |
- |
PREDICTED: uncharacterized protein LOC105647419 [Jatropha curcas] |
| 7 |
Hb_000014_170 |
0.0918620892 |
- |
- |
hypothetical protein CISIN_1g0379742mg, partial [Citrus sinensis] |
| 8 |
Hb_003126_040 |
0.0981972255 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000042_210 |
0.1014411751 |
- |
- |
PREDICTED: uncharacterized protein LOC105632808 [Jatropha curcas] |
| 10 |
Hb_010368_020 |
0.1031675416 |
- |
- |
PREDICTED: uncharacterized protein LOC105630301 [Jatropha curcas] |
| 11 |
Hb_007007_050 |
0.1079945589 |
- |
- |
PREDICTED: ubiquitin-related modifier 1 homolog 2 [Malus domestica] |
| 12 |
Hb_005016_090 |
0.1083717649 |
- |
- |
PREDICTED: uncharacterized protein LOC105642243 [Jatropha curcas] |
| 13 |
Hb_000390_130 |
0.1108046135 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B isoform X2 [Jatropha curcas] |
| 14 |
Hb_004976_020 |
0.1108374134 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 15 |
Hb_003086_070 |
0.1108903127 |
- |
- |
PREDICTED: iron-sulfur cluster co-chaperone protein HscB, mitochondrial [Jatropha curcas] |
| 16 |
Hb_003849_060 |
0.1110547021 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000920_280 |
0.1130369278 |
- |
- |
Protein SENSITIVITY TO RED LIGHT REDUCED, putative [Ricinus communis] |
| 18 |
Hb_001329_130 |
0.1146844089 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_021422_010 |
0.11613913 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 1, mitochondrial [Jatropha curcas] |
| 20 |
Hb_028900_010 |
0.1175402619 |
- |
- |
PREDICTED: uncharacterized protein LOC105631404 isoform X2 [Jatropha curcas] |