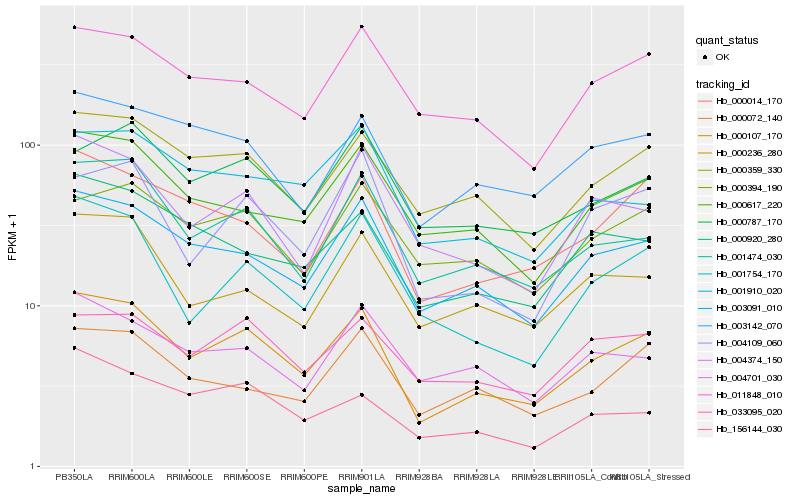
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000107_170 |
0.0 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC25 homolog [Jatropha curcas] |
| 2 |
Hb_003091_010 |
0.0856804012 |
- |
- |
PREDICTED: uncharacterized protein LOC105131887 [Populus euphratica] |
| 3 |
Hb_000359_330 |
0.0944228792 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001474_030 |
0.0949007854 |
- |
- |
PREDICTED: uncharacterized protein LOC105650916 [Jatropha curcas] |
| 5 |
Hb_000014_170 |
0.1019728951 |
- |
- |
hypothetical protein CISIN_1g0379742mg, partial [Citrus sinensis] |
| 6 |
Hb_000617_220 |
0.1084421524 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 7 |
Hb_001754_170 |
0.1090747514 |
- |
- |
PREDICTED: 60S ribosome subunit biogenesis protein NIP7 homolog [Jatropha curcas] |
| 8 |
Hb_011848_010 |
0.1103964764 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 9 |
Hb_003142_070 |
0.1104259961 |
- |
- |
hypothetical protein F383_14657 [Gossypium arboreum] |
| 10 |
Hb_004374_150 |
0.1134455986 |
- |
- |
PREDICTED: uncharacterized protein LOC105650290 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000920_280 |
0.1144280377 |
- |
- |
Protein SENSITIVITY TO RED LIGHT REDUCED, putative [Ricinus communis] |
| 12 |
Hb_004109_060 |
0.1145958807 |
- |
- |
hypothetical protein POPTR_0019s12520g [Populus trichocarpa] |
| 13 |
Hb_033095_020 |
0.1163250225 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000236_280 |
0.1178461434 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 22 homolog isoform X1 [Jatropha curcas] |
| 15 |
Hb_000072_140 |
0.1180865566 |
- |
- |
- |
| 16 |
Hb_000787_170 |
0.1192106068 |
- |
- |
hypothetical protein POPTR_0005s06400g [Populus trichocarpa] |
| 17 |
Hb_001910_020 |
0.1207870811 |
- |
- |
PREDICTED: ubiquitin-associated domain-containing protein 2 [Jatropha curcas] |
| 18 |
Hb_156144_030 |
0.1228030493 |
- |
- |
hypothetical protein RCOM_0187400 [Ricinus communis] |
| 19 |
Hb_000394_190 |
0.1241497424 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_004701_030 |
0.124242375 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |