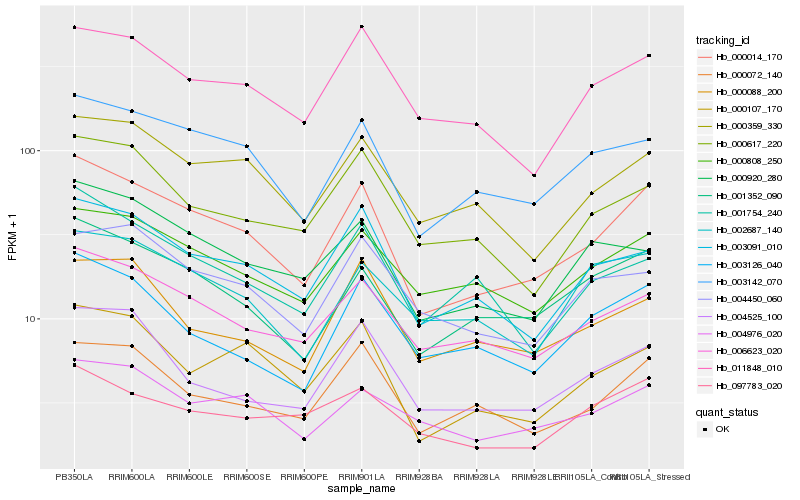
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000014_170 |
0.0 |
- |
- |
hypothetical protein CISIN_1g0379742mg, partial [Citrus sinensis] |
| 2 |
Hb_003142_070 |
0.0883359074 |
- |
- |
hypothetical protein F383_14657 [Gossypium arboreum] |
| 3 |
Hb_001352_090 |
0.0918620892 |
- |
- |
PREDICTED: UPF0161 protein At3g09310 [Jatropha curcas] |
| 4 |
Hb_006623_020 |
0.0959069705 |
- |
- |
PREDICTED: protein LURP-one-related 7 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003091_010 |
0.0966216349 |
- |
- |
PREDICTED: uncharacterized protein LOC105131887 [Populus euphratica] |
| 6 |
Hb_000072_140 |
0.100535006 |
- |
- |
- |
| 7 |
Hb_000107_170 |
0.1019728951 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC25 homolog [Jatropha curcas] |
| 8 |
Hb_000359_330 |
0.1032131774 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000808_250 |
0.1071322222 |
- |
- |
PREDICTED: uncharacterized protein LOC105647419 [Jatropha curcas] |
| 10 |
Hb_011848_010 |
0.1076322523 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 11 |
Hb_004976_020 |
0.1107608674 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 12 |
Hb_000920_280 |
0.1115177821 |
- |
- |
Protein SENSITIVITY TO RED LIGHT REDUCED, putative [Ricinus communis] |
| 13 |
Hb_004525_100 |
0.112066171 |
- |
- |
hypothetical protein JCGZ_18418 [Jatropha curcas] |
| 14 |
Hb_003126_040 |
0.112601235 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000617_220 |
0.1143362098 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 16 |
Hb_001754_240 |
0.1175234086 |
- |
- |
PREDICTED: mitochondrial thiamine pyrophosphate carrier isoform X1 [Jatropha curcas] |
| 17 |
Hb_002687_140 |
0.1192254975 |
- |
- |
hypothetical protein JCGZ_24463 [Jatropha curcas] |
| 18 |
Hb_097783_020 |
0.1194112902 |
- |
- |
PREDICTED: small G protein signaling modulator 1 [Jatropha curcas] |
| 19 |
Hb_004450_060 |
0.1195689889 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000088_200 |
0.1206831135 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |