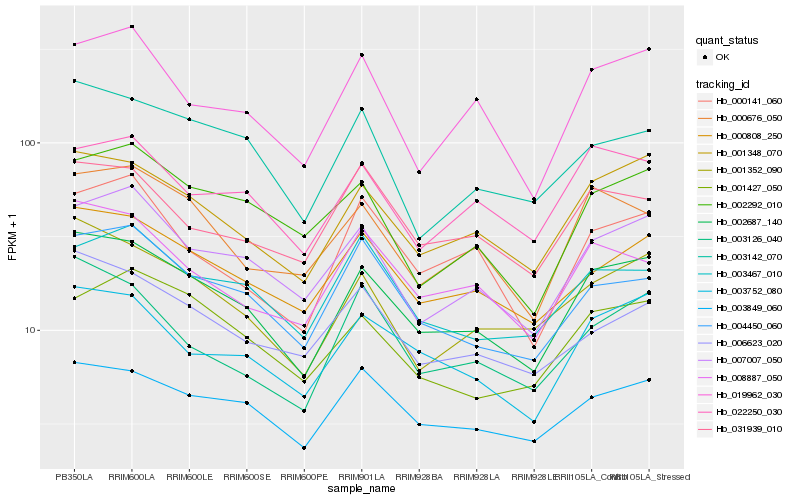
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002687_140 |
0.0 |
- |
- |
hypothetical protein JCGZ_24463 [Jatropha curcas] |
| 2 |
Hb_001348_070 |
0.0562343074 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 3 |
Hb_003752_080 |
0.0752112249 |
- |
- |
PREDICTED: ubiquitin-like-conjugating enzyme ATG10 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000808_250 |
0.076075758 |
- |
- |
PREDICTED: uncharacterized protein LOC105647419 [Jatropha curcas] |
| 5 |
Hb_003849_060 |
0.0772840464 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 6 |
Hb_007007_050 |
0.0783901748 |
- |
- |
PREDICTED: ubiquitin-related modifier 1 homolog 2 [Malus domestica] |
| 7 |
Hb_001352_090 |
0.0800430672 |
- |
- |
PREDICTED: UPF0161 protein At3g09310 [Jatropha curcas] |
| 8 |
Hb_008887_050 |
0.0891453904 |
- |
- |
PREDICTED: DNA-binding protein BIN4 [Jatropha curcas] |
| 9 |
Hb_000676_050 |
0.0894692261 |
- |
- |
PREDICTED: thioredoxin domain-containing protein PLP3B-like [Gossypium raimondii] |
| 10 |
Hb_004450_060 |
0.0908600022 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X2 [Jatropha curcas] |
| 11 |
Hb_019962_030 |
0.0917694663 |
- |
- |
PREDICTED: uncharacterized protein LOC105650622 [Jatropha curcas] |
| 12 |
Hb_003142_070 |
0.0922807045 |
- |
- |
hypothetical protein F383_14657 [Gossypium arboreum] |
| 13 |
Hb_006623_020 |
0.0947376455 |
- |
- |
PREDICTED: protein LURP-one-related 7 isoform X1 [Jatropha curcas] |
| 14 |
Hb_022250_030 |
0.0951958736 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002292_010 |
0.0956460765 |
- |
- |
hypothetical protein CICLE_v10029628mg [Citrus clementina] |
| 16 |
Hb_001427_050 |
0.096058007 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 17 |
Hb_003126_040 |
0.0985913039 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000141_060 |
0.0991956265 |
- |
- |
PREDICTED: uncharacterized protein LOC105632248 [Jatropha curcas] |
| 19 |
Hb_031939_010 |
0.1007395497 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 20 |
Hb_003467_010 |
0.1007397664 |
- |
- |
PREDICTED: probable thiol methyltransferase 2 [Jatropha curcas] |