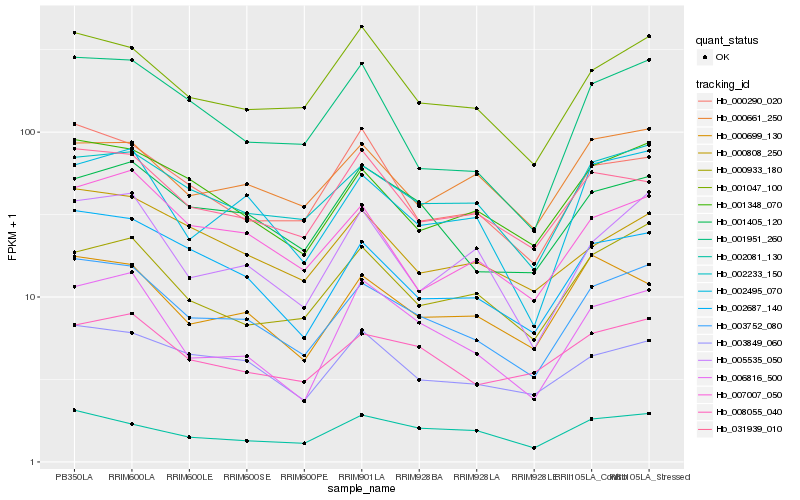
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003752_080 |
0.0 |
- |
- |
PREDICTED: ubiquitin-like-conjugating enzyme ATG10 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002233_150 |
0.0736467497 |
- |
- |
PREDICTED: uncharacterized protein LOC105633093 [Jatropha curcas] |
| 3 |
Hb_002687_140 |
0.0752112249 |
- |
- |
hypothetical protein JCGZ_24463 [Jatropha curcas] |
| 4 |
Hb_001348_070 |
0.0757472168 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 5 |
Hb_001047_100 |
0.0877177699 |
- |
- |
40S ribosomal protein S18, putative [Ricinus communis] |
| 6 |
Hb_002495_070 |
0.0902345536 |
- |
- |
PREDICTED: uncharacterized protein LOC105634731 [Jatropha curcas] |
| 7 |
Hb_001405_120 |
0.0903986361 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit H [Jatropha curcas] |
| 8 |
Hb_003849_060 |
0.0908474567 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 9 |
Hb_008055_040 |
0.0918954201 |
- |
- |
PREDICTED: chromatin assembly factor 1 subunit FAS2 [Jatropha curcas] |
| 10 |
Hb_000933_180 |
0.0967169722 |
- |
- |
PREDICTED: chorismate mutase 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000808_250 |
0.0979887182 |
- |
- |
PREDICTED: uncharacterized protein LOC105647419 [Jatropha curcas] |
| 12 |
Hb_031939_010 |
0.0981460974 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 13 |
Hb_007007_050 |
0.0993473276 |
- |
- |
PREDICTED: ubiquitin-related modifier 1 homolog 2 [Malus domestica] |
| 14 |
Hb_000699_130 |
0.0998054377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002081_130 |
0.0998944042 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 16 |
Hb_006816_500 |
0.1003003409 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 17 |
Hb_005535_050 |
0.1016170938 |
- |
- |
PREDICTED: uncharacterized protein At2g39795, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_000661_250 |
0.1016628099 |
- |
- |
PREDICTED: RNA-binding protein 8A [Jatropha curcas] |
| 19 |
Hb_000290_020 |
0.1018216923 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit K-like isoform X2 [Populus euphratica] |
| 20 |
Hb_001951_260 |
0.1019749782 |
- |
- |
60S ribosomal protein L32, putative [Ricinus communis] |