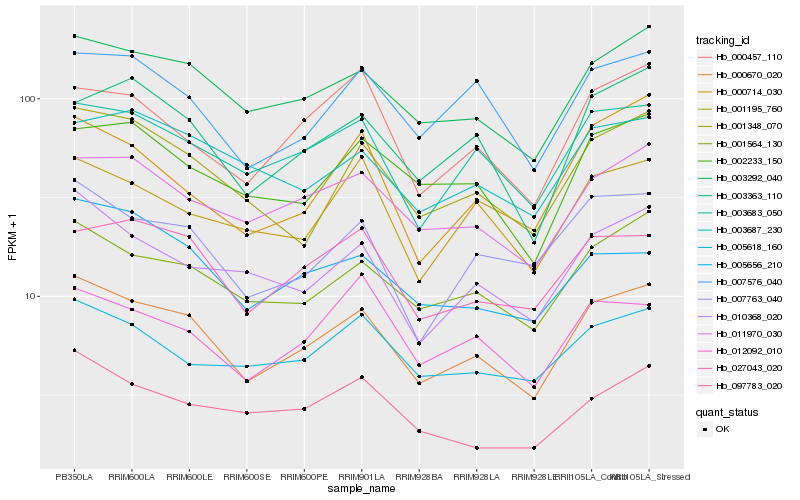
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000670_020 |
0.0 |
- |
- |
PREDICTED: chromatin modification-related protein EAF3-like [Jatropha curcas] |
| 2 |
Hb_003292_040 |
0.0628676147 |
transcription factor |
TF Family: MBF1 |
uncharacterized protein LOC100500420 [Glycine max] |
| 3 |
Hb_001564_130 |
0.0699277721 |
- |
- |
PREDICTED: single-stranded DNA-binding protein, mitochondrial isoform X1 [Jatropha curcas] |
| 4 |
Hb_003683_050 |
0.077745808 |
- |
- |
PREDICTED: uncharacterized protein LOC105650619 [Jatropha curcas] |
| 5 |
Hb_001348_070 |
0.0797553262 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 6 |
Hb_011970_030 |
0.0834631276 |
- |
- |
PREDICTED: uncharacterized protein LOC105641805 [Jatropha curcas] |
| 7 |
Hb_010368_020 |
0.083821467 |
- |
- |
PREDICTED: uncharacterized protein LOC105630301 [Jatropha curcas] |
| 8 |
Hb_007763_040 |
0.0853677937 |
- |
- |
- |
| 9 |
Hb_027043_020 |
0.0876796947 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003363_110 |
0.0878177479 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_007576_040 |
0.0881754156 |
- |
- |
PREDICTED: uncharacterized protein LOC105169221 [Sesamum indicum] |
| 12 |
Hb_002233_150 |
0.089824145 |
- |
- |
PREDICTED: uncharacterized protein LOC105633093 [Jatropha curcas] |
| 13 |
Hb_001195_760 |
0.0942763606 |
- |
- |
PREDICTED: 30S ribosomal protein S9, chloroplastic-like [Populus euphratica] |
| 14 |
Hb_005618_160 |
0.0944403518 |
- |
- |
PREDICTED: putative proline--tRNA ligase C19C7.06 [Jatropha curcas] |
| 15 |
Hb_000457_110 |
0.0965751627 |
- |
- |
Mitochondrial ribosomal protein L37 isoform 1 [Theobroma cacao] |
| 16 |
Hb_000714_030 |
0.0972548593 |
- |
- |
Nucleosome assembly [Gossypium arboreum] |
| 17 |
Hb_003687_230 |
0.0982928229 |
- |
- |
PREDICTED: actin-depolymerizing factor-like [Jatropha curcas] |
| 18 |
Hb_097783_020 |
0.0999653956 |
- |
- |
PREDICTED: small G protein signaling modulator 1 [Jatropha curcas] |
| 19 |
Hb_005656_210 |
0.1001972125 |
- |
- |
PREDICTED: uncharacterized protein LOC105637440 [Jatropha curcas] |
| 20 |
Hb_012092_010 |
0.1001972225 |
- |
- |
PREDICTED: uncharacterized protein LOC105635929 [Jatropha curcas] |