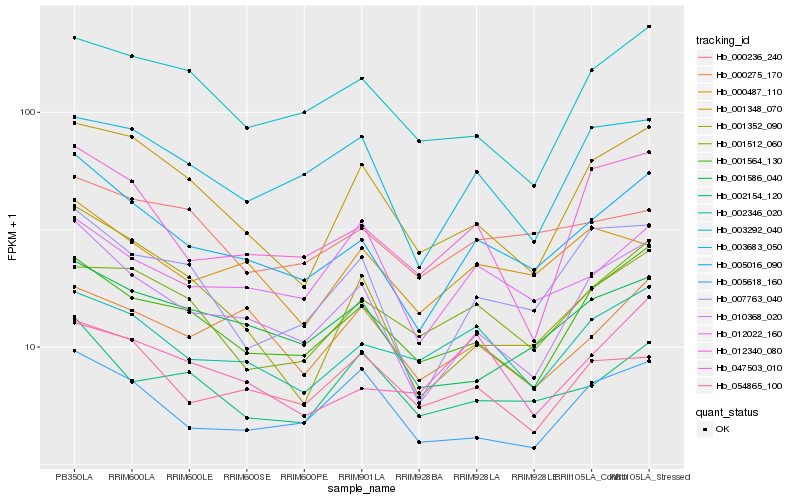
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005016_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642243 [Jatropha curcas] |
| 2 |
Hb_010368_020 |
0.0653569253 |
- |
- |
PREDICTED: uncharacterized protein LOC105630301 [Jatropha curcas] |
| 3 |
Hb_001564_130 |
0.0863610466 |
- |
- |
PREDICTED: single-stranded DNA-binding protein, mitochondrial isoform X1 [Jatropha curcas] |
| 4 |
Hb_007763_040 |
0.0931064545 |
- |
- |
- |
| 5 |
Hb_002346_020 |
0.0947427566 |
- |
- |
- |
| 6 |
Hb_001586_040 |
0.0970119966 |
- |
- |
hypothetical protein JCGZ_17187 [Jatropha curcas] |
| 7 |
Hb_003683_050 |
0.1019779157 |
- |
- |
PREDICTED: uncharacterized protein LOC105650619 [Jatropha curcas] |
| 8 |
Hb_001348_070 |
0.103219118 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_002154_120 |
0.103578441 |
- |
- |
PREDICTED: uncharacterized protein LOC105630563 [Jatropha curcas] |
| 10 |
Hb_000487_110 |
0.1038345477 |
- |
- |
PREDICTED: cyclin-T1-3-like [Jatropha curcas] |
| 11 |
Hb_005618_160 |
0.1040370728 |
- |
- |
PREDICTED: putative proline--tRNA ligase C19C7.06 [Jatropha curcas] |
| 12 |
Hb_012340_080 |
0.1052041141 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 13 |
Hb_001512_060 |
0.1060681016 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g35850, mitochondrial [Jatropha curcas] |
| 14 |
Hb_012022_160 |
0.1061175201 |
- |
- |
PREDICTED: uncharacterized protein LOC105646602 [Jatropha curcas] |
| 15 |
Hb_054865_100 |
0.107637164 |
- |
- |
PREDICTED: probable protein S-acyltransferase 12 isoform X1 [Jatropha curcas] |
| 16 |
Hb_047503_010 |
0.1082051707 |
- |
- |
PREDICTED: putative protein N-methyltransferase YNL024C [Jatropha curcas] |
| 17 |
Hb_001352_090 |
0.1083717649 |
- |
- |
PREDICTED: UPF0161 protein At3g09310 [Jatropha curcas] |
| 18 |
Hb_003292_040 |
0.1087677061 |
transcription factor |
TF Family: MBF1 |
uncharacterized protein LOC100500420 [Glycine max] |
| 19 |
Hb_000236_240 |
0.1090338371 |
- |
- |
Pre-mRNA-splicing factor cwc15, putative [Ricinus communis] |
| 20 |
Hb_000275_170 |
0.110458955 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b-like [Jatropha curcas] |