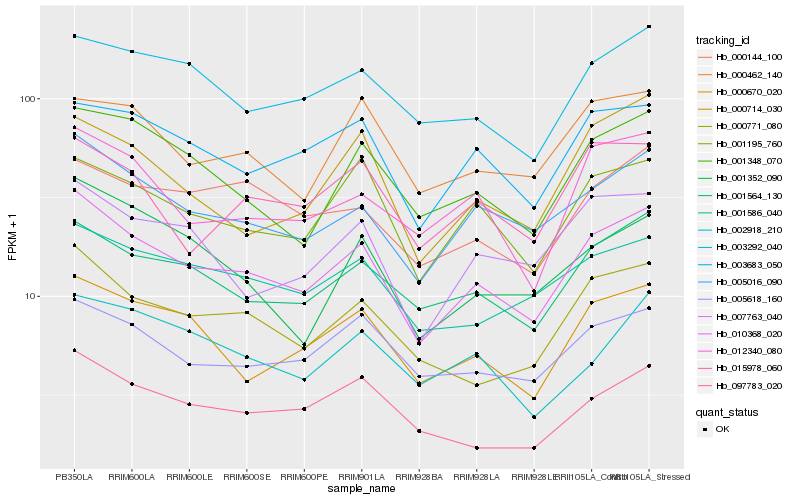
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010368_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630301 [Jatropha curcas] |
| 2 |
Hb_005016_090 |
0.0653569253 |
- |
- |
PREDICTED: uncharacterized protein LOC105642243 [Jatropha curcas] |
| 3 |
Hb_001564_130 |
0.0756583971 |
- |
- |
PREDICTED: single-stranded DNA-binding protein, mitochondrial isoform X1 [Jatropha curcas] |
| 4 |
Hb_000771_080 |
0.0806896991 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 5 |
Hb_000670_020 |
0.083821467 |
- |
- |
PREDICTED: chromatin modification-related protein EAF3-like [Jatropha curcas] |
| 6 |
Hb_001348_070 |
0.0876602702 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 7 |
Hb_001195_760 |
0.0878250458 |
- |
- |
PREDICTED: 30S ribosomal protein S9, chloroplastic-like [Populus euphratica] |
| 8 |
Hb_005618_160 |
0.0880453934 |
- |
- |
PREDICTED: putative proline--tRNA ligase C19C7.06 [Jatropha curcas] |
| 9 |
Hb_003683_050 |
0.0885774608 |
- |
- |
PREDICTED: uncharacterized protein LOC105650619 [Jatropha curcas] |
| 10 |
Hb_012340_080 |
0.0903274575 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 11 |
Hb_001586_040 |
0.0908958608 |
- |
- |
hypothetical protein JCGZ_17187 [Jatropha curcas] |
| 12 |
Hb_007763_040 |
0.0915130535 |
- |
- |
- |
| 13 |
Hb_003292_040 |
0.0921511483 |
transcription factor |
TF Family: MBF1 |
uncharacterized protein LOC100500420 [Glycine max] |
| 14 |
Hb_097783_020 |
0.09432865 |
- |
- |
PREDICTED: small G protein signaling modulator 1 [Jatropha curcas] |
| 15 |
Hb_000144_100 |
0.0991355845 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA-like 2, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000714_030 |
0.100562742 |
- |
- |
Nucleosome assembly [Gossypium arboreum] |
| 17 |
Hb_000462_140 |
0.1012009669 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 28 [Jatropha curcas] |
| 18 |
Hb_015978_060 |
0.1023043255 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_001352_090 |
0.1031675416 |
- |
- |
PREDICTED: UPF0161 protein At3g09310 [Jatropha curcas] |
| 20 |
Hb_002918_210 |
0.1052243146 |
- |
- |
PREDICTED: uncharacterized protein LOC105138897 isoform X2 [Populus euphratica] |