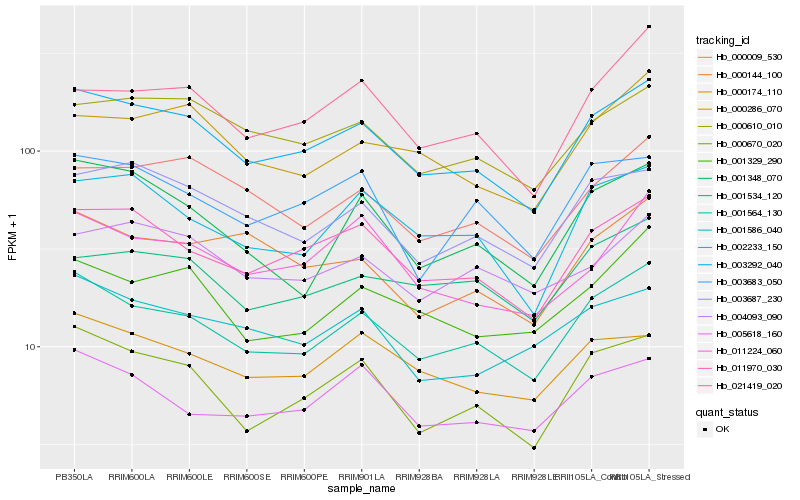
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003292_040 |
0.0 |
transcription factor |
TF Family: MBF1 |
uncharacterized protein LOC100500420 [Glycine max] |
| 2 |
Hb_001564_130 |
0.0482783205 |
- |
- |
PREDICTED: single-stranded DNA-binding protein, mitochondrial isoform X1 [Jatropha curcas] |
| 3 |
Hb_011970_030 |
0.0494751684 |
- |
- |
PREDICTED: uncharacterized protein LOC105641805 [Jatropha curcas] |
| 4 |
Hb_000670_020 |
0.0628676147 |
- |
- |
PREDICTED: chromatin modification-related protein EAF3-like [Jatropha curcas] |
| 5 |
Hb_000610_010 |
0.0671910204 |
- |
- |
PREDICTED: SKP1-like protein 1B [Musa acuminata subsp. malaccensis] |
| 6 |
Hb_003687_230 |
0.0678669168 |
- |
- |
PREDICTED: actin-depolymerizing factor-like [Jatropha curcas] |
| 7 |
Hb_002233_150 |
0.0711860582 |
- |
- |
PREDICTED: uncharacterized protein LOC105633093 [Jatropha curcas] |
| 8 |
Hb_000009_530 |
0.0749775329 |
- |
- |
PREDICTED: uncharacterized protein LOC105639628 [Jatropha curcas] |
| 9 |
Hb_000286_070 |
0.0774320033 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 10 |
Hb_000174_110 |
0.0793999241 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 11 |
Hb_011224_060 |
0.0805899989 |
- |
- |
hypothetical protein POPTR_0011s16880g [Populus trichocarpa] |
| 12 |
Hb_001329_290 |
0.081111799 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 13 |
Hb_003683_050 |
0.0823888159 |
- |
- |
PREDICTED: uncharacterized protein LOC105650619 [Jatropha curcas] |
| 14 |
Hb_021419_020 |
0.0828791024 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 15 |
Hb_001534_120 |
0.0843377343 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 37 homolog 1 [Gossypium raimondii] |
| 16 |
Hb_004093_090 |
0.0846080428 |
- |
- |
PREDICTED: uncharacterized protein LOC105633550 [Jatropha curcas] |
| 17 |
Hb_005618_160 |
0.0848085657 |
- |
- |
PREDICTED: putative proline--tRNA ligase C19C7.06 [Jatropha curcas] |
| 18 |
Hb_001586_040 |
0.0848123616 |
- |
- |
hypothetical protein JCGZ_17187 [Jatropha curcas] |
| 19 |
Hb_001348_070 |
0.085582387 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_000144_100 |
0.0861495337 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA-like 2, mitochondrial [Jatropha curcas] |