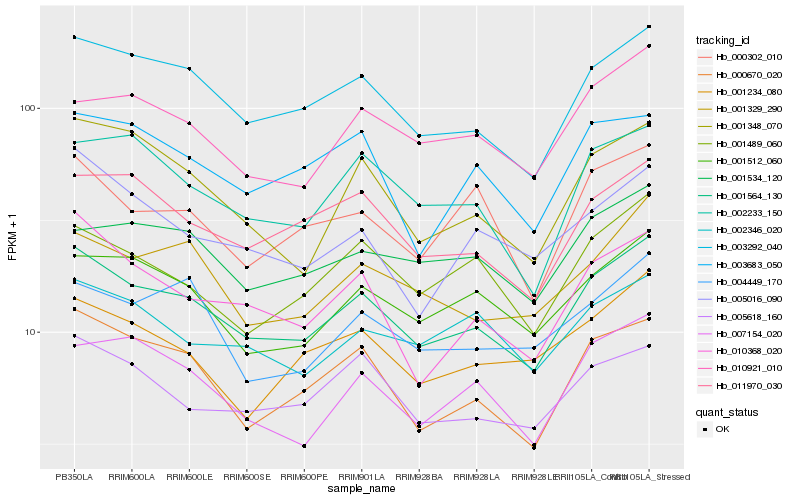
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001564_130 |
0.0 |
- |
- |
PREDICTED: single-stranded DNA-binding protein, mitochondrial isoform X1 [Jatropha curcas] |
| 2 |
Hb_003292_040 |
0.0482783205 |
transcription factor |
TF Family: MBF1 |
uncharacterized protein LOC100500420 [Glycine max] |
| 3 |
Hb_000670_020 |
0.0699277721 |
- |
- |
PREDICTED: chromatin modification-related protein EAF3-like [Jatropha curcas] |
| 4 |
Hb_001512_060 |
0.0736165502 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g35850, mitochondrial [Jatropha curcas] |
| 5 |
Hb_011970_030 |
0.0747760094 |
- |
- |
PREDICTED: uncharacterized protein LOC105641805 [Jatropha curcas] |
| 6 |
Hb_010368_020 |
0.0756583971 |
- |
- |
PREDICTED: uncharacterized protein LOC105630301 [Jatropha curcas] |
| 7 |
Hb_001348_070 |
0.0766432044 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 8 |
Hb_001329_290 |
0.0770993642 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 9 |
Hb_002233_150 |
0.078306716 |
- |
- |
PREDICTED: uncharacterized protein LOC105633093 [Jatropha curcas] |
| 10 |
Hb_001489_060 |
0.0785929229 |
- |
- |
PREDICTED: uncharacterized protein LOC105649778 isoform X2 [Jatropha curcas] |
| 11 |
Hb_005618_160 |
0.0792419771 |
- |
- |
PREDICTED: putative proline--tRNA ligase C19C7.06 [Jatropha curcas] |
| 12 |
Hb_010921_010 |
0.0808531887 |
- |
- |
Small nuclear ribonucleoprotein family protein [Theobroma cacao] |
| 13 |
Hb_007154_020 |
0.0814002783 |
- |
- |
- |
| 14 |
Hb_002346_020 |
0.0839002833 |
- |
- |
- |
| 15 |
Hb_001534_120 |
0.0844200753 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 37 homolog 1 [Gossypium raimondii] |
| 16 |
Hb_000302_010 |
0.084914812 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |
| 17 |
Hb_005016_090 |
0.0863610466 |
- |
- |
PREDICTED: uncharacterized protein LOC105642243 [Jatropha curcas] |
| 18 |
Hb_001234_080 |
0.0872804461 |
- |
- |
PREDICTED: eyes absent homolog 2-like isoform X2 [Musa acuminata subsp. malaccensis] |
| 19 |
Hb_004449_170 |
0.0877701672 |
- |
- |
PREDICTED: uncharacterized protein LOC105631285 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003683_050 |
0.0881767403 |
- |
- |
PREDICTED: uncharacterized protein LOC105650619 [Jatropha curcas] |