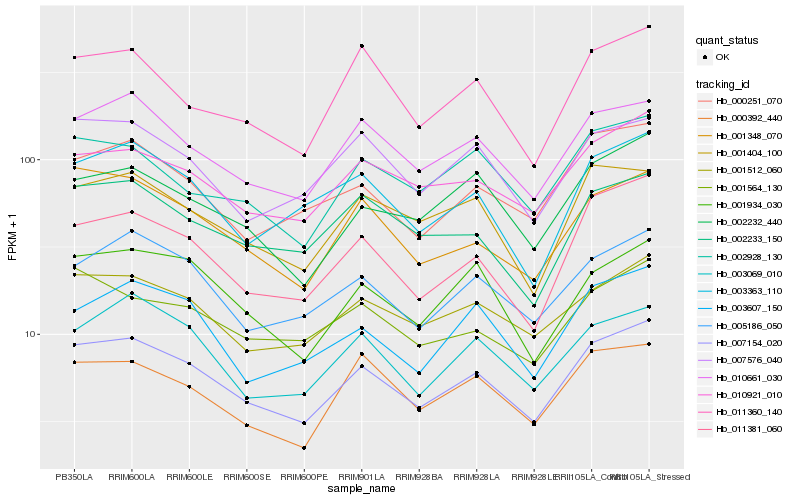
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007154_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_011381_060 |
0.0637533702 |
- |
- |
PREDICTED: ADP-ribosylation factor 3 [Jatropha curcas] |
| 3 |
Hb_010661_030 |
0.0663054499 |
- |
- |
hypothetical protein PHAVU_009G146200g [Phaseolus vulgaris] |
| 4 |
Hb_001512_060 |
0.0684492598 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g35850, mitochondrial [Jatropha curcas] |
| 5 |
Hb_005186_050 |
0.068838714 |
- |
- |
PREDICTED: uncharacterized protein LOC105628509 [Jatropha curcas] |
| 6 |
Hb_001348_070 |
0.0692127385 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 7 |
Hb_010921_010 |
0.0705715201 |
- |
- |
Small nuclear ribonucleoprotein family protein [Theobroma cacao] |
| 8 |
Hb_000251_070 |
0.0746281544 |
- |
- |
hypothetical protein L484_010641 [Morus notabilis] |
| 9 |
Hb_001934_030 |
0.0759771994 |
transcription factor |
TF Family: HSF |
Heat shock factor protein, putative [Ricinus communis] |
| 10 |
Hb_002232_440 |
0.0763546143 |
- |
- |
PREDICTED: uncharacterized protein LOC105130257 [Populus euphratica] |
| 11 |
Hb_003607_150 |
0.0764069734 |
- |
- |
PREDICTED: thioredoxin-like protein 4B [Jatropha curcas] |
| 12 |
Hb_003363_110 |
0.0767265484 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_001404_100 |
0.0790730684 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit K [Gossypium raimondii] |
| 14 |
Hb_003069_010 |
0.079340535 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 2-like [Jatropha curcas] |
| 15 |
Hb_002233_150 |
0.0801939114 |
- |
- |
PREDICTED: uncharacterized protein LOC105633093 [Jatropha curcas] |
| 16 |
Hb_000392_440 |
0.0811614273 |
- |
- |
PREDICTED: nuclear ribonuclease Z isoform X1 [Jatropha curcas] |
| 17 |
Hb_007576_040 |
0.0812371131 |
- |
- |
PREDICTED: uncharacterized protein LOC105169221 [Sesamum indicum] |
| 18 |
Hb_011360_140 |
0.0813437403 |
- |
- |
40S ribosomal protein S19, putative [Ricinus communis] |
| 19 |
Hb_001564_130 |
0.0814002783 |
- |
- |
PREDICTED: single-stranded DNA-binding protein, mitochondrial isoform X1 [Jatropha curcas] |
| 20 |
Hb_002928_130 |
0.0828791722 |
- |
- |
PREDICTED: small nuclear ribonucleoprotein Sm D3 [Jatropha curcas] |