| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
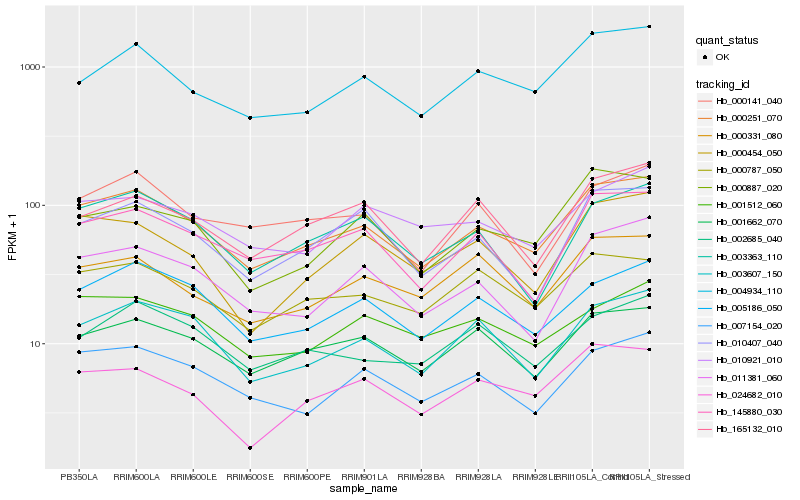
Hb_000251_070 |
0.0 |
- |
- |
hypothetical protein L484_010641 [Morus notabilis] |
| 2 |
Hb_005186_050 |
0.072902865 |
- |
- |
PREDICTED: uncharacterized protein LOC105628509 [Jatropha curcas] |
| 3 |
Hb_004934_110 |
0.0736271992 |
transcription factor |
TF Family: HMG |
hypothetical protein B456_002G115100 [Gossypium raimondii] |
| 4 |
Hb_001662_070 |
0.0736655033 |
- |
- |
PREDICTED: thiol-disulfide oxidoreductase LTO1 [Jatropha curcas] |
| 5 |
Hb_145880_030 |
0.0737910687 |
- |
- |
ubiquitin-conjugating enzyme E2-25kD, putative [Ricinus communis] |
| 6 |
Hb_003607_150 |
0.0741231691 |
- |
- |
PREDICTED: thioredoxin-like protein 4B [Jatropha curcas] |
| 7 |
Hb_007154_020 |
0.0746281544 |
- |
- |
- |
| 8 |
Hb_011381_060 |
0.0782370401 |
- |
- |
PREDICTED: ADP-ribosylation factor 3 [Jatropha curcas] |
| 9 |
Hb_000787_050 |
0.082964711 |
- |
- |
PREDICTED: electron transfer flavoprotein subunit alpha, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_010407_040 |
0.0835111706 |
- |
- |
PREDICTED: uncharacterized protein At4g14342 isoform X1 [Nelumbo nucifera] |
| 11 |
Hb_003363_110 |
0.0839974163 |
- |
- |
protein with unknown function [Ricinus communis] |
| 12 |
Hb_000887_020 |
0.0864296529 |
- |
- |
PREDICTED: uncharacterized protein LOC105633527 [Jatropha curcas] |
| 13 |
Hb_024682_010 |
0.088588305 |
- |
- |
sterol isomerase, putative [Ricinus communis] |
| 14 |
Hb_010921_010 |
0.088652357 |
- |
- |
Small nuclear ribonucleoprotein family protein [Theobroma cacao] |
| 15 |
Hb_000141_040 |
0.0891380321 |
- |
- |
hypothetical protein CICLE_v10022858mg [Citrus clementina] |
| 16 |
Hb_165132_010 |
0.0894083625 |
- |
- |
PREDICTED: uncharacterized protein LOC105134496 [Populus euphratica] |
| 17 |
Hb_001512_060 |
0.0899234236 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g35850, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000454_050 |
0.0900542921 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9 [Vitis vinifera] |
| 19 |
Hb_002685_040 |
0.0907635021 |
- |
- |
hypothetical protein CICLE_v10017252mg [Citrus clementina] |
| 20 |
Hb_000331_080 |
0.0907658754 |
- |
- |
PREDICTED: UPF0235 protein At5g63440 isoform X1 [Gossypium raimondii] |