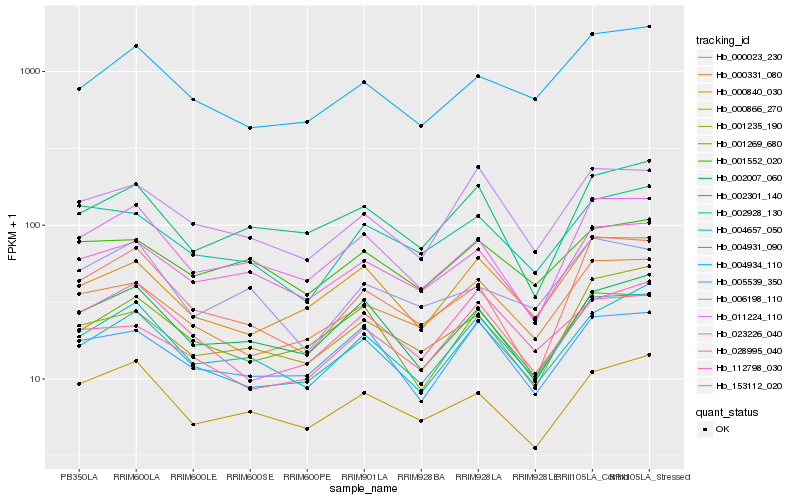
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004657_050 |
0.0 |
- |
- |
PREDICTED: tRNA-dihydrouridine(16/17) synthase [NAD(P)(+)]-like [Jatropha curcas] |
| 2 |
Hb_023226_040 |
0.0634665826 |
- |
- |
PREDICTED: kxDL motif-containing protein 1 [Jatropha curcas] |
| 3 |
Hb_028995_040 |
0.0642687227 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 11 isoform X1 [Jatropha curcas] |
| 4 |
Hb_011224_110 |
0.0646702563 |
- |
- |
Pre-mRNA-splicing factor cwc15, putative [Ricinus communis] |
| 5 |
Hb_002007_060 |
0.06491632 |
- |
- |
hypothetical protein JCGZ_23323 [Jatropha curcas] |
| 6 |
Hb_004931_090 |
0.069208228 |
- |
- |
PREDICTED: chromatin modification-related protein MEAF6-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_000866_270 |
0.0732152716 |
- |
- |
PREDICTED: uncharacterized protein LOC105642993 [Jatropha curcas] |
| 8 |
Hb_000331_080 |
0.076202032 |
- |
- |
PREDICTED: UPF0235 protein At5g63440 isoform X1 [Gossypium raimondii] |
| 9 |
Hb_004934_110 |
0.0762656064 |
transcription factor |
TF Family: HMG |
hypothetical protein B456_002G115100 [Gossypium raimondii] |
| 10 |
Hb_000840_030 |
0.0773220877 |
transcription factor |
TF Family: IWS1 |
PREDICTED: probable mediator of RNA polymerase II transcription subunit 26c isoform X2 [Jatropha curcas] |
| 11 |
Hb_002301_140 |
0.0785468895 |
- |
- |
PREDICTED: uncharacterized protein At4g28440 [Jatropha curcas] |
| 12 |
Hb_005539_350 |
0.079678631 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001269_680 |
0.0821817703 |
- |
- |
PREDICTED: uncharacterized protein LOC105132670 isoform X2 [Populus euphratica] |
| 14 |
Hb_001552_020 |
0.0839210326 |
- |
- |
PREDICTED: uncharacterized protein LOC105639653 [Jatropha curcas] |
| 15 |
Hb_001235_190 |
0.0847036839 |
- |
- |
PREDICTED: peroxisome biogenesis protein 19-2-like [Jatropha curcas] |
| 16 |
Hb_000023_230 |
0.0851189704 |
- |
- |
PREDICTED: uncharacterized protein LOC105637526 [Jatropha curcas] |
| 17 |
Hb_006198_110 |
0.0869632768 |
- |
- |
PREDICTED: uncharacterized protein At4g13200, chloroplastic [Jatropha curcas] |
| 18 |
Hb_153112_020 |
0.0869982024 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 22b-like [Jatropha curcas] |
| 19 |
Hb_002928_130 |
0.0873249909 |
- |
- |
PREDICTED: small nuclear ribonucleoprotein Sm D3 [Jatropha curcas] |
| 20 |
Hb_112798_030 |
0.0873471908 |
- |
- |
conserved hypothetical protein [Ricinus communis] |