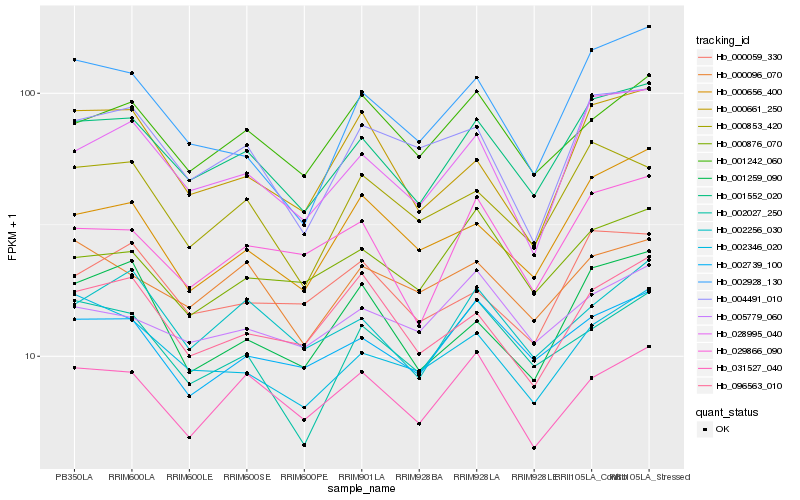
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001552_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639653 [Jatropha curcas] |
| 2 |
Hb_028995_040 |
0.0530587008 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 11 isoform X1 [Jatropha curcas] |
| 3 |
Hb_029866_090 |
0.0663103127 |
- |
- |
PREDICTED: uncharacterized protein LOC105648377 [Jatropha curcas] |
| 4 |
Hb_002739_100 |
0.066795681 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000656_400 |
0.0680537686 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 9 [Jatropha curcas] |
| 6 |
Hb_002256_030 |
0.0686645431 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002346_020 |
0.0690387733 |
- |
- |
- |
| 8 |
Hb_000853_420 |
0.0693342164 |
- |
- |
PREDICTED: phosphoribosylformylglycinamidine cyclo-ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 9 |
Hb_004491_010 |
0.0723463905 |
- |
- |
60S ribosomal protein L51, putative [Ricinus communis] |
| 10 |
Hb_001242_060 |
0.072424937 |
- |
- |
PREDICTED: uncharacterized protein LOC105139508 [Populus euphratica] |
| 11 |
Hb_031527_040 |
0.0728715965 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001259_090 |
0.0731409189 |
- |
- |
hypothetical protein B456_005G056700 [Gossypium raimondii] |
| 13 |
Hb_000059_330 |
0.0734923912 |
- |
- |
PREDICTED: polyadenylate-binding protein 1 [Jatropha curcas] |
| 14 |
Hb_000096_070 |
0.0750753778 |
- |
- |
PREDICTED: ruvB-like 2 [Jatropha curcas] |
| 15 |
Hb_002928_130 |
0.0751049706 |
- |
- |
PREDICTED: small nuclear ribonucleoprotein Sm D3 [Jatropha curcas] |
| 16 |
Hb_096563_010 |
0.075606084 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Prunus mume] |
| 17 |
Hb_000661_250 |
0.076340086 |
- |
- |
PREDICTED: RNA-binding protein 8A [Jatropha curcas] |
| 18 |
Hb_002027_250 |
0.0766271071 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000876_070 |
0.0777342582 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 20 |
Hb_005779_060 |
0.0794931784 |
- |
- |
PREDICTED: uncharacterized protein LOC105643212 isoform X2 [Jatropha curcas] |