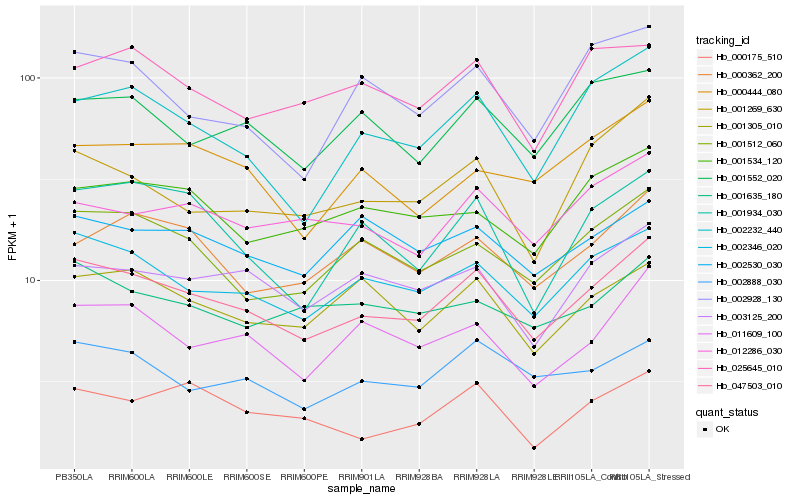
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_047503_010 |
0.0 |
- |
- |
PREDICTED: putative protein N-methyltransferase YNL024C [Jatropha curcas] |
| 2 |
Hb_002346_020 |
0.0573824286 |
- |
- |
- |
| 3 |
Hb_001512_060 |
0.0803135407 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g35850, mitochondrial [Jatropha curcas] |
| 4 |
Hb_011609_100 |
0.0842293587 |
- |
- |
PREDICTED: 30S ribosomal protein 3, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_002888_030 |
0.0844511023 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 6 |
Hb_002232_440 |
0.0857987608 |
- |
- |
PREDICTED: uncharacterized protein LOC105130257 [Populus euphratica] |
| 7 |
Hb_003125_200 |
0.0867137244 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 30 [Jatropha curcas] |
| 8 |
Hb_001552_020 |
0.0902333055 |
- |
- |
PREDICTED: uncharacterized protein LOC105639653 [Jatropha curcas] |
| 9 |
Hb_001305_010 |
0.0906392095 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001635_180 |
0.0927088442 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At5g53840-like [Jatropha curcas] |
| 11 |
Hb_012286_030 |
0.0930974471 |
transcription factor |
TF Family: MED7 |
PREDICTED: mediator of RNA polymerase II transcription subunit 7a [Solanum lycopersicum] |
| 12 |
Hb_001534_120 |
0.0938781553 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 37 homolog 1 [Gossypium raimondii] |
| 13 |
Hb_000362_200 |
0.0950521851 |
- |
- |
exonuclease family protein [Populus trichocarpa] |
| 14 |
Hb_001934_030 |
0.0951618886 |
transcription factor |
TF Family: HSF |
Heat shock factor protein, putative [Ricinus communis] |
| 15 |
Hb_001269_630 |
0.0954158507 |
- |
- |
PREDICTED: uncharacterized protein LOC105630301 [Jatropha curcas] |
| 16 |
Hb_002530_030 |
0.0962801326 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 4 [Jatropha curcas] |
| 17 |
Hb_025645_010 |
0.0967203215 |
- |
- |
PREDICTED: proteasome subunit beta type-5 [Jatropha curcas] |
| 18 |
Hb_002928_130 |
0.0969057288 |
- |
- |
PREDICTED: small nuclear ribonucleoprotein Sm D3 [Jatropha curcas] |
| 19 |
Hb_000175_510 |
0.097042519 |
- |
- |
PREDICTED: probable DNA replication complex GINS protein PSF1 [Jatropha curcas] |
| 20 |
Hb_000444_080 |
0.0971349208 |
- |
- |
hypothetical protein POPTR_0019s10500g [Populus trichocarpa] |