| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
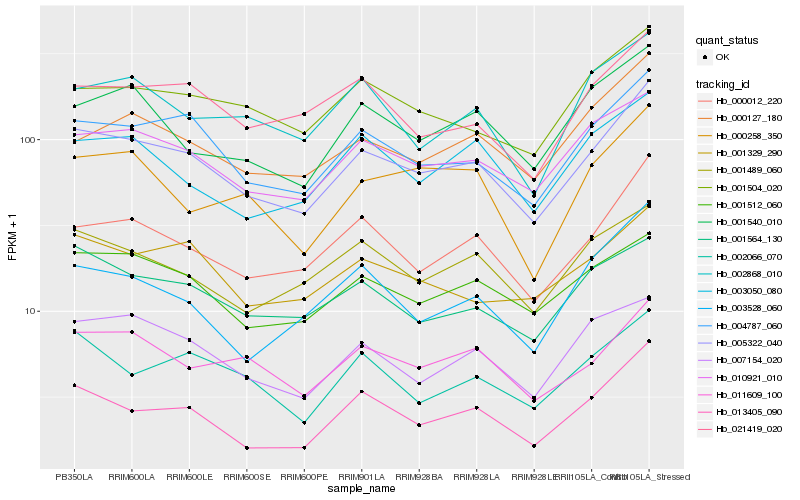
Hb_005322_040 |
0.0 |
- |
- |
60S ribosomal protein L9, putative [Ricinus communis] |
| 2 |
Hb_004787_060 |
0.059088888 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 3 |
Hb_000012_220 |
0.0678259282 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_010921_010 |
0.0731595586 |
- |
- |
Small nuclear ribonucleoprotein family protein [Theobroma cacao] |
| 5 |
Hb_013405_090 |
0.082087405 |
- |
- |
PREDICTED: uncharacterized protein LOC105648433 isoform X1 [Jatropha curcas] |
| 6 |
Hb_011609_100 |
0.0822810103 |
- |
- |
PREDICTED: 30S ribosomal protein 3, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_001512_060 |
0.0855351921 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g35850, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000127_180 |
0.0877031244 |
- |
- |
40S ribosomal protein S18, putative [Ricinus communis] |
| 9 |
Hb_001504_020 |
0.0887609973 |
- |
- |
40S ribosomal protein S12A [Hevea brasiliensis] |
| 10 |
Hb_001329_290 |
0.0890186074 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 11 |
Hb_021419_020 |
0.0893383531 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 12 |
Hb_001489_060 |
0.0901205188 |
- |
- |
PREDICTED: uncharacterized protein LOC105649778 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001564_130 |
0.0911775572 |
- |
- |
PREDICTED: single-stranded DNA-binding protein, mitochondrial isoform X1 [Jatropha curcas] |
| 14 |
Hb_007154_020 |
0.0915716134 |
- |
- |
- |
| 15 |
Hb_003050_080 |
0.0923335175 |
- |
- |
40S ribosomal protein S14, putative [Ricinus communis] |
| 16 |
Hb_001540_010 |
0.0927354884 |
- |
- |
PREDICTED: uncharacterized protein LOC105639920 [Jatropha curcas] |
| 17 |
Hb_002066_070 |
0.0968958677 |
- |
- |
PREDICTED: uncharacterized protein LOC105631043 [Jatropha curcas] |
| 18 |
Hb_000258_350 |
0.0975244108 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 19 |
Hb_002868_010 |
0.0982366722 |
- |
- |
60S ribosomal protein L30, putative [Ricinus communis] |
| 20 |
Hb_003528_060 |
0.0983556689 |
- |
- |
PREDICTED: uncharacterized protein LOC105638256 isoform X2 [Jatropha curcas] |