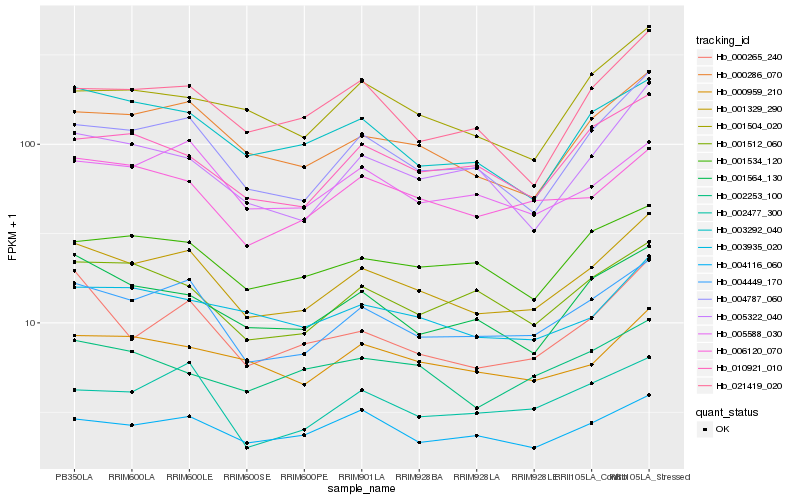
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001329_290 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 2 |
Hb_004449_170 |
0.0436292093 |
- |
- |
PREDICTED: uncharacterized protein LOC105631285 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000286_070 |
0.0585582128 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 4 |
Hb_004787_060 |
0.0624758106 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 5 |
Hb_001564_130 |
0.0770993642 |
- |
- |
PREDICTED: single-stranded DNA-binding protein, mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_003292_040 |
0.081111799 |
transcription factor |
TF Family: MBF1 |
uncharacterized protein LOC100500420 [Glycine max] |
| 7 |
Hb_000265_240 |
0.0815708839 |
- |
- |
PREDICTED: uncharacterized protein LOC105643590 isoform X1 [Jatropha curcas] |
| 8 |
Hb_006120_070 |
0.082261901 |
- |
- |
PREDICTED: ER membrane protein complex subunit 6 [Jatropha curcas] |
| 9 |
Hb_003935_020 |
0.0830310554 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_001534_120 |
0.0831448775 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 37 homolog 1 [Gossypium raimondii] |
| 11 |
Hb_010921_010 |
0.0845288017 |
- |
- |
Small nuclear ribonucleoprotein family protein [Theobroma cacao] |
| 12 |
Hb_000959_210 |
0.0850933489 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 27 [Jatropha curcas] |
| 13 |
Hb_005588_030 |
0.0851668379 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Eucalyptus grandis] |
| 14 |
Hb_001512_060 |
0.0854134637 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g35850, mitochondrial [Jatropha curcas] |
| 15 |
Hb_002477_300 |
0.0876477616 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_004116_060 |
0.0881591552 |
- |
- |
PREDICTED: nuclear pore complex protein NUP88 [Jatropha curcas] |
| 17 |
Hb_005322_040 |
0.0890186074 |
- |
- |
60S ribosomal protein L9, putative [Ricinus communis] |
| 18 |
Hb_021419_020 |
0.0897428733 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 19 |
Hb_001504_020 |
0.0931881703 |
- |
- |
40S ribosomal protein S12A [Hevea brasiliensis] |
| 20 |
Hb_002253_100 |
0.0948729787 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |