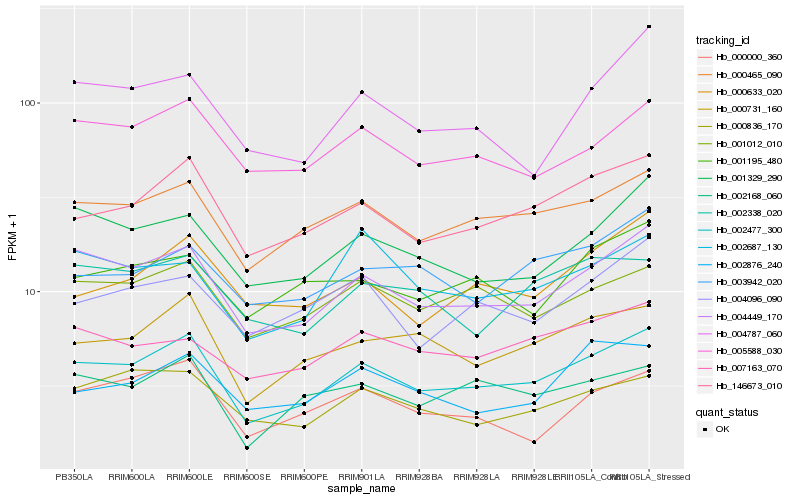
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002477_300 |
0.0 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_004449_170 |
0.0544723817 |
- |
- |
PREDICTED: uncharacterized protein LOC105631285 isoform X1 [Jatropha curcas] |
| 3 |
Hb_146673_010 |
0.0656833962 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 4 |
Hb_000465_090 |
0.0683649219 |
- |
- |
PREDICTED: uncharacterized protein LOC105632289 [Jatropha curcas] |
| 5 |
Hb_000731_160 |
0.0777281077 |
- |
- |
PREDICTED: adenosine deaminase-like protein [Jatropha curcas] |
| 6 |
Hb_001329_290 |
0.0876477616 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 7 |
Hb_001012_010 |
0.0879135205 |
- |
- |
PREDICTED: XIAP-associated factor 1 [Jatropha curcas] |
| 8 |
Hb_005588_030 |
0.0882451762 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Eucalyptus grandis] |
| 9 |
Hb_002168_060 |
0.0914946628 |
- |
- |
PREDICTED: inositol-pentakisphosphate 2-kinase-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000836_170 |
0.0956080573 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 11 |
Hb_000633_020 |
0.0976588258 |
- |
- |
PREDICTED: uncharacterized protein LOC105630002 [Jatropha curcas] |
| 12 |
Hb_004096_090 |
0.0986955354 |
- |
- |
Plastid-specific 30S ribosomal protein 3, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_007163_070 |
0.10040325 |
- |
- |
PREDICTED: tRNA-dihydrouridine(20) synthase [NAD(P)+]-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_003942_020 |
0.1005065527 |
- |
- |
PREDICTED: multiple RNA-binding domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001195_480 |
0.1011763981 |
- |
- |
cop9 complex subunit, putative [Ricinus communis] |
| 16 |
Hb_000000_360 |
0.1016808998 |
- |
- |
PREDICTED: methyltransferase-like protein 2 [Populus euphratica] |
| 17 |
Hb_002338_020 |
0.1020775063 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR34A [Jatropha curcas] |
| 18 |
Hb_004787_060 |
0.1021038669 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 19 |
Hb_002876_240 |
0.1024964366 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_002687_130 |
0.1025926055 |
- |
- |
PREDICTED: uncharacterized protein LOC105631153 isoform X1 [Jatropha curcas] |