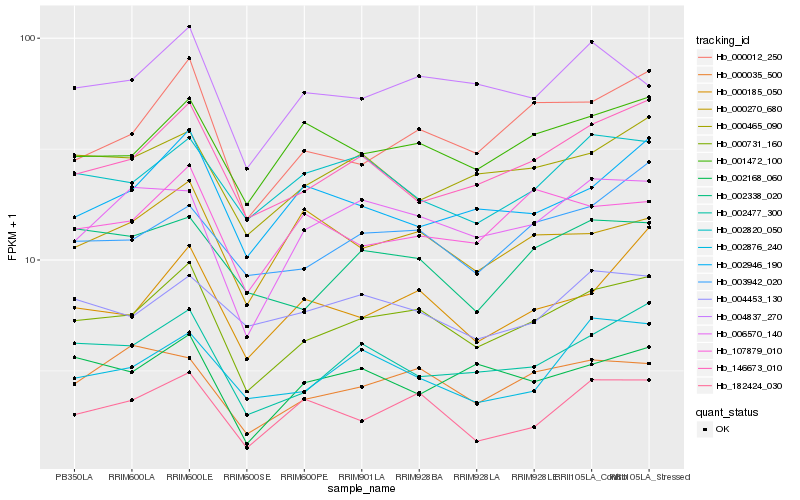
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000731_160 |
0.0 |
- |
- |
PREDICTED: adenosine deaminase-like protein [Jatropha curcas] |
| 2 |
Hb_000012_250 |
0.0763711962 |
- |
- |
hypothetical protein L484_019524 [Morus notabilis] |
| 3 |
Hb_002477_300 |
0.0777281077 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_146673_010 |
0.0799581087 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 5 |
Hb_001472_100 |
0.086302349 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 6 |
Hb_006570_140 |
0.090691266 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 7 |
Hb_000185_050 |
0.0912391229 |
- |
- |
PREDICTED: FKBP12-interacting protein of 37 kDa [Jatropha curcas] |
| 8 |
Hb_000465_090 |
0.0921997952 |
- |
- |
PREDICTED: uncharacterized protein LOC105632289 [Jatropha curcas] |
| 9 |
Hb_182424_030 |
0.0928278414 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 8 [Vitis vinifera] |
| 10 |
Hb_003942_020 |
0.0928646074 |
- |
- |
PREDICTED: multiple RNA-binding domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004837_270 |
0.0940465554 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1 [Jatropha curcas] |
| 12 |
Hb_000035_500 |
0.0973192765 |
- |
- |
PREDICTED: ADP-ribosylation factor 1 isoform X2 [Tarenaya hassleriana] |
| 13 |
Hb_002946_190 |
0.0975116172 |
- |
- |
PREDICTED: nudix hydrolase 26, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002820_050 |
0.0980067514 |
- |
- |
PREDICTED: putative glutathione-specific gamma-glutamylcyclotransferase 2 [Jatropha curcas] |
| 15 |
Hb_002338_020 |
0.0986575817 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR34A [Jatropha curcas] |
| 16 |
Hb_107879_010 |
0.1000425961 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 17 |
Hb_002876_240 |
0.1016322919 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_000270_680 |
0.1024661675 |
- |
- |
hypothetical protein POPTR_0015s08420g [Populus trichocarpa] |
| 19 |
Hb_002168_060 |
0.1032544391 |
- |
- |
PREDICTED: inositol-pentakisphosphate 2-kinase-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_004453_130 |
0.1032872055 |
- |
- |
PREDICTED: tRNA (cytosine-5-)-methyltransferase isoform X1 [Jatropha curcas] |