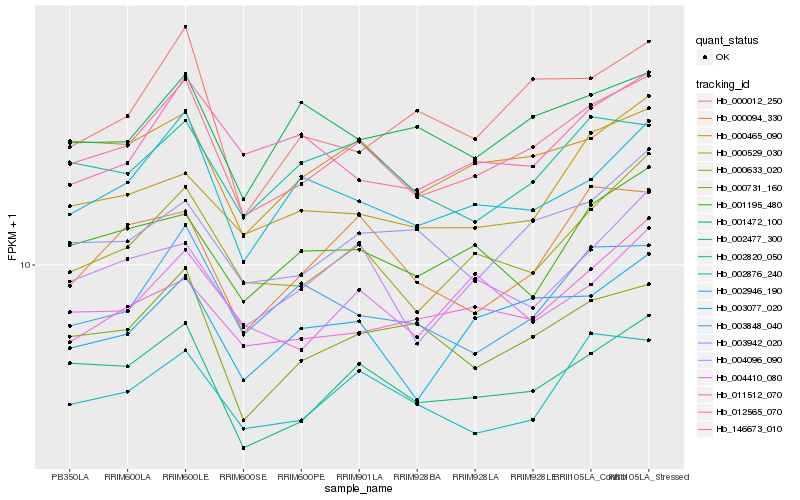
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_146673_010 |
0.0 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 2 |
Hb_000633_020 |
0.0656604826 |
- |
- |
PREDICTED: uncharacterized protein LOC105630002 [Jatropha curcas] |
| 3 |
Hb_002477_300 |
0.0656833962 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_000465_090 |
0.0708788829 |
- |
- |
PREDICTED: uncharacterized protein LOC105632289 [Jatropha curcas] |
| 5 |
Hb_002946_190 |
0.0785988595 |
- |
- |
PREDICTED: nudix hydrolase 26, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003077_020 |
0.078684928 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_000731_160 |
0.0799581087 |
- |
- |
PREDICTED: adenosine deaminase-like protein [Jatropha curcas] |
| 8 |
Hb_003942_020 |
0.0844027456 |
- |
- |
PREDICTED: multiple RNA-binding domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002876_240 |
0.085456078 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_002820_050 |
0.08739026 |
- |
- |
PREDICTED: putative glutathione-specific gamma-glutamylcyclotransferase 2 [Jatropha curcas] |
| 11 |
Hb_003848_040 |
0.0905512118 |
- |
- |
PREDICTED: phosphatidylinositol-glycan biosynthesis class X protein isoform X1 [Jatropha curcas] |
| 12 |
Hb_004096_090 |
0.0913040144 |
- |
- |
Plastid-specific 30S ribosomal protein 3, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_000094_330 |
0.0915471003 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 4 [Jatropha curcas] |
| 14 |
Hb_000529_030 |
0.091651359 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_000012_250 |
0.0917682534 |
- |
- |
hypothetical protein L484_019524 [Morus notabilis] |
| 16 |
Hb_004410_080 |
0.0924504821 |
- |
- |
cellular nucleic acid binding protein, putative [Ricinus communis] |
| 17 |
Hb_001195_480 |
0.0937424155 |
- |
- |
cop9 complex subunit, putative [Ricinus communis] |
| 18 |
Hb_011512_070 |
0.0951861508 |
transcription factor |
TF Family: PHD |
Inhibitor of growth protein, putative [Ricinus communis] |
| 19 |
Hb_001472_100 |
0.0963867829 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 20 |
Hb_012565_070 |
0.0966658589 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |