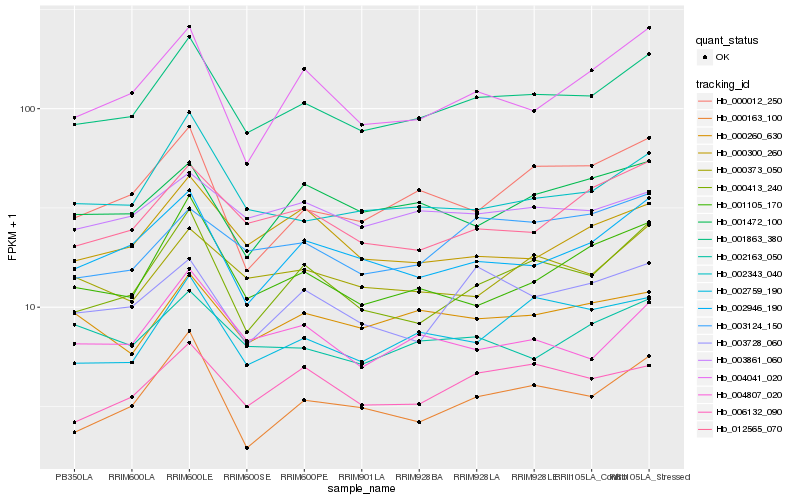
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001863_380 |
0.0 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 2 |
Hb_000413_240 |
0.0686439637 |
- |
- |
PREDICTED: transcription factor ILR3-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_002946_190 |
0.0690976472 |
- |
- |
PREDICTED: nudix hydrolase 26, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002343_040 |
0.0702245015 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_012565_070 |
0.0757306662 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 6 |
Hb_000373_050 |
0.0770010529 |
- |
- |
PREDICTED: protein WHI3 [Jatropha curcas] |
| 7 |
Hb_002759_190 |
0.0782631612 |
- |
- |
PREDICTED: protein NLRC3 [Jatropha curcas] |
| 8 |
Hb_001105_170 |
0.0786035562 |
- |
- |
PREDICTED: uncharacterized protein LOC105642727 isoform X1 [Jatropha curcas] |
| 9 |
Hb_006132_090 |
0.0796987755 |
- |
- |
PREDICTED: origin of replication complex subunit 5 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004807_020 |
0.0801422228 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 14b-like [Jatropha curcas] |
| 11 |
Hb_004041_020 |
0.0818634301 |
- |
- |
ATP synthase D chain, mitochondrial, putative [Ricinus communis] |
| 12 |
Hb_000163_100 |
0.0832462481 |
- |
- |
PREDICTED: uncharacterized protein LOC105642537 [Jatropha curcas] |
| 13 |
Hb_002163_050 |
0.0839885963 |
- |
- |
hypothetical protein POPTR_0014s15490g [Populus trichocarpa] |
| 14 |
Hb_000260_630 |
0.0842979248 |
- |
- |
PREDICTED: altered inheritance rate of mitochondria protein 25 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003861_060 |
0.0843007712 |
- |
- |
PREDICTED: treacle protein [Jatropha curcas] |
| 16 |
Hb_000300_260 |
0.0844193323 |
- |
- |
PREDICTED: replication factor C subunit 3 [Jatropha curcas] |
| 17 |
Hb_000012_250 |
0.0851989069 |
- |
- |
hypothetical protein L484_019524 [Morus notabilis] |
| 18 |
Hb_003728_060 |
0.0859037425 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 4-like [Jatropha curcas] |
| 19 |
Hb_003124_150 |
0.0883642284 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 28-like [Jatropha curcas] |
| 20 |
Hb_001472_100 |
0.0886881213 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |